Parallel Squared Technology Institute (PTI)
@ParallelSqTech
PTI is a nonprofit focused research organization scaling proteomic technologies to explore proteome biology. A member of the @SchmidtFutures & @Convergent_FROs.
Bạn có thể thích
This community is building foundational capabilities for technological progress. Its nonprofit initiatives allow broadly sharing these capabilities. I am happy to support this mission 🚀
We opened a new Head of Development position to join PTI's mission of creating a sustainable and scalable model for non-profit research. We would love to hear from you if you are the right fit for our team! jobs.lever.co/convergentrese…
And, we are back with another presentation from this year's Research Fest. PTI's Senior Computational Scientist, Kevin McDonnell, talks about developing joint modeling (aka JMod) of mass spectra for empowering multiplexed DIA proteomics. youtube.com/watch?v=plK4TC…
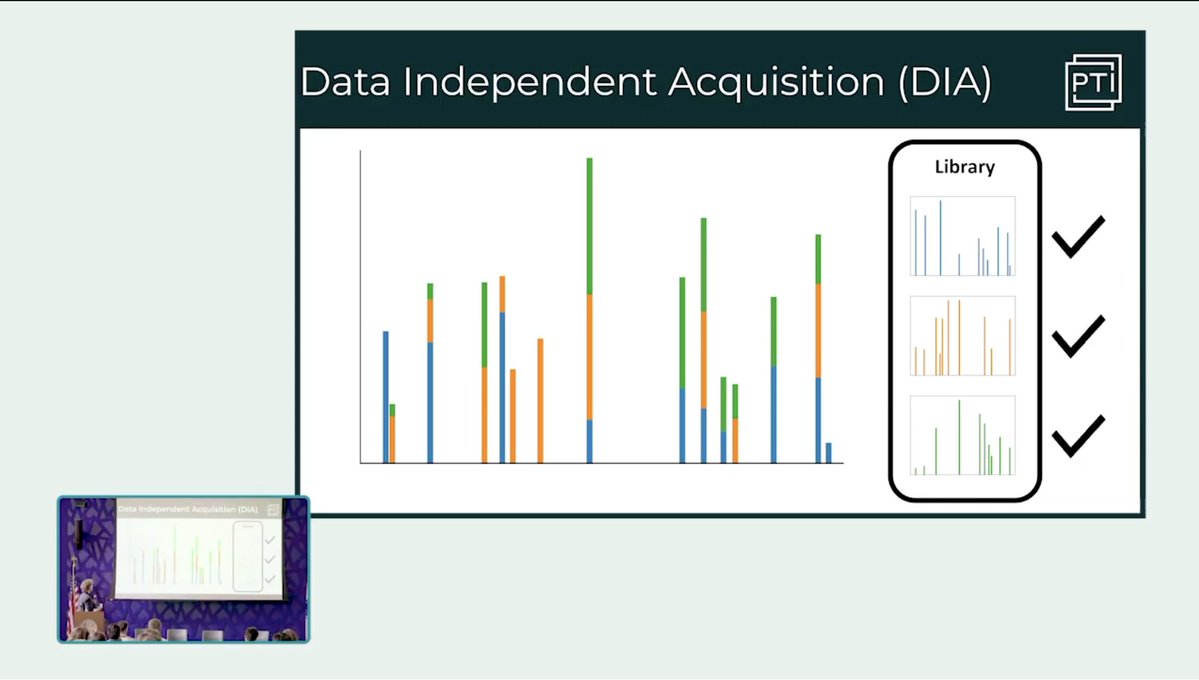
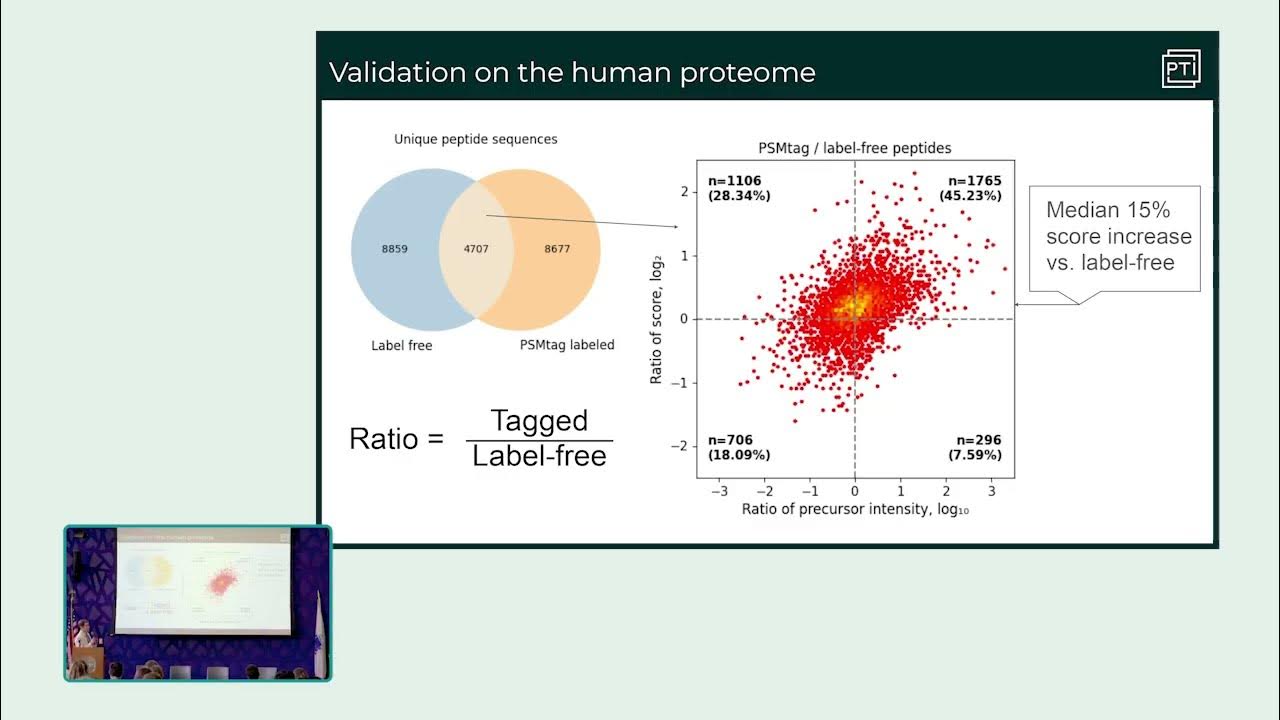
Throwback Thursday continues with a presentation by Maddy Yeh and Mark Adamo from Research Fest: "Tags for improving peptide sequencing and throughput in sensitive proteomics". Watch on YouTube: youtube.com/watch?v=jA2I9x…

Throwback Thursday continues with a presentation by Maddy Yeh and Mark Adamo: "Tags for improving peptide sequencing and throughput in sensitive proteomics" from Research Fest. Watch on YouTube: youtube.com/watch?v=jA2I9x…
youtube.com
YouTube
Tags for improving peptide sequencing and throughput in sensitive...
The counting statistics for single-cell MS support accurate estimates of protein abundance in single cells, which is essential for quantitative analysis. Further, measuring and modeling protein degradation is key to biological interpretations. pubs.acs.org/doi/10.1021/ac…
Earlier this year, @ParallelSqTech hosted a research fest. We began with a broad perspective: ◾️ A century of remarkable progress! Proteomics drove conceptual discoveries and medical treatments. Listen to what's next! Proteomics: The arc of progress youtu.be/Sc2QLXb84XI?si…

It's good to look back and see how far we have come since Research Fest took place almost six months ago. Over the next few weeks, we will post some throwback videos from the presentations. Let's start with opening remarks by PTI's Director, @slavov_n: youtu.be/Sc2QLXb84XI

youtube.com
YouTube
Proteomics: The arc of progress | Nikolai Slavov
Our large scale measurements of protein functions and conformations can help realize the potential of AI. When developing AI models, we emphasize careful validation & rigorous benchmarking.
In this article, I shared my perspective on AI. AI excels at supervised learning, and well annotated datasets can help unlock new AI capabilities. 🔗 nature.com/articles/s4225… OA 🔗 rdcu.be/eI4JW
Interpreting single-cell measurements is a challenging and promising frontier ! Solid interpretations begin with the experimental design and considerations about the information that the measurements can provide.
mRNA-mRNA correlations across the single cells from a tissue are rarely interpreted. They differ from the corresponding protein-protein correlations? 𝐖𝐡𝐲 ❓ biorxiv.org/content/10.110… 🧵

𝐏𝐫𝐨𝐭𝐞𝐚𝐬𝐨𝐦𝐞𝐬 𝐩𝐥𝐚𝐲 𝐤𝐞𝐲 𝐫𝐨𝐥𝐞𝐬 𝐢𝐧 𝐧𝐞𝐮𝐫𝐨𝐝𝐞𝐠𝐞𝐧𝐞𝐫𝐚𝐭𝐢𝐨𝐧 Neuronal inflammation induces PSMB8 Integrating PSMB8 into neuronal proteasomes reduces their activity This causes PFKFB3 accumulation, a metabolic switch, and ferroptosis 🧵

I am looking forward to ICSB next week. I will present: 1⃣ New technology from @ParallelSqTech: parallelsq.org/blog/proteomic… 2⃣ New biology from @slavovLab: biorxiv.org/content/10.110…

Protein abundance is regulated by many mechanisms. We quantified them across the single cells in a mammalian tissue: 1⃣ The results revealed simple rules ! 2⃣ They may alter how you interpret single-cell DNA(RNA)-sed data. ⚠️ See @_AndrewLeduc's 🧵 biorxiv.org/content/10.110…

The big one is finally out!! In this paper, we set out to provide insight into the fundamental question; How do the individual cells from complex tissues regulate their proteomes? Brief summary of our findings 👇 biorxiv.org/content/10.110…
Staggering sample loading to fit more samples per mass spec run: biorxiv.org/content/10.110…
Screening for more peptide barcodes: biorxiv.org/content/10.110…
A clear summary of our approach & progress towards scaling up (single-cell) proteomics.
I was super excited to learn from @NikoMcCarty's essay, "How to Scale Proteomics", that there's an FRO working on high-throughput single-cell proteomics (@ParallelSqTech), and even more excited to see the progress they've made in just 2 years via their preprints. So here's a…
I was super excited to learn from @NikoMcCarty's essay, "How to Scale Proteomics", that there's an FRO working on high-throughput single-cell proteomics (@ParallelSqTech), and even more excited to see the progress they've made in just 2 years via their preprints. So here's a…
PTI is growing and we are looking for more scientists to join our team! Neurodegenerative Disease Senior Scientist: bit.ly/4n35ND7 Computational Biologist: bit.ly/4gplQsJ Research Chemist: bit.ly/46n0Q13
United States Xu hướng
- 1. Texans 37K posts
- 2. World Series 107K posts
- 3. CJ Stroud 6,594 posts
- 4. Mariners 91.1K posts
- 5. Blue Jays 93.3K posts
- 6. Seahawks 34.9K posts
- 7. Seattle 51.2K posts
- 8. Springer 65.9K posts
- 9. Nick Caley 2,493 posts
- 10. Dan Wilson 4,242 posts
- 11. #WWERaw 59.2K posts
- 12. White House 302K posts
- 13. Nico Collins 2,105 posts
- 14. Kenneth Walker 2,518 posts
- 15. Demeco 1,739 posts
- 16. Bazardo 3,101 posts
- 17. Sam Darnold 4,220 posts
- 18. Baker 37.2K posts
- 19. LA Knight 7,796 posts
- 20. Munoz 10.3K posts
Bạn có thể thích
-
 Slavov Laboratory
Slavov Laboratory
@slavovLab -
 Prof. Nikolai Slavov
Prof. Nikolai Slavov
@slavov_n -
 Single-Cell Proteomics Conference
Single-Cell Proteomics Conference
@SCP_meeting -
 Human Proteome
Human Proteome
@hupo_org -
 CoxLab
CoxLab
@TheCoxLab -
 JesperOlsenLab
JesperOlsenLab
@JesperOlsenLab -
 Molecular & Cellular Proteomics
Molecular & Cellular Proteomics
@molcellprot -
 MaxQuant
MaxQuant
@maxquant -
 E11 Bio
E11 Bio
@E11BIO -
 Biognosys
Biognosys
@biognosys -
 Evosep
Evosep
@EvosepBio -
 Dysautonomia Intl.
Dysautonomia Intl.
@Dysautonomia -
 The Sato Lab (Kei Sato)
The Sato Lab (Kei Sato)
@SystemsVirology -
 Buck Institute
Buck Institute
@BuckInstitute -
 PreOmics
PreOmics
@PreOmics
Something went wrong.
Something went wrong.