#spatialtranscriptomics ผลการค้นหา
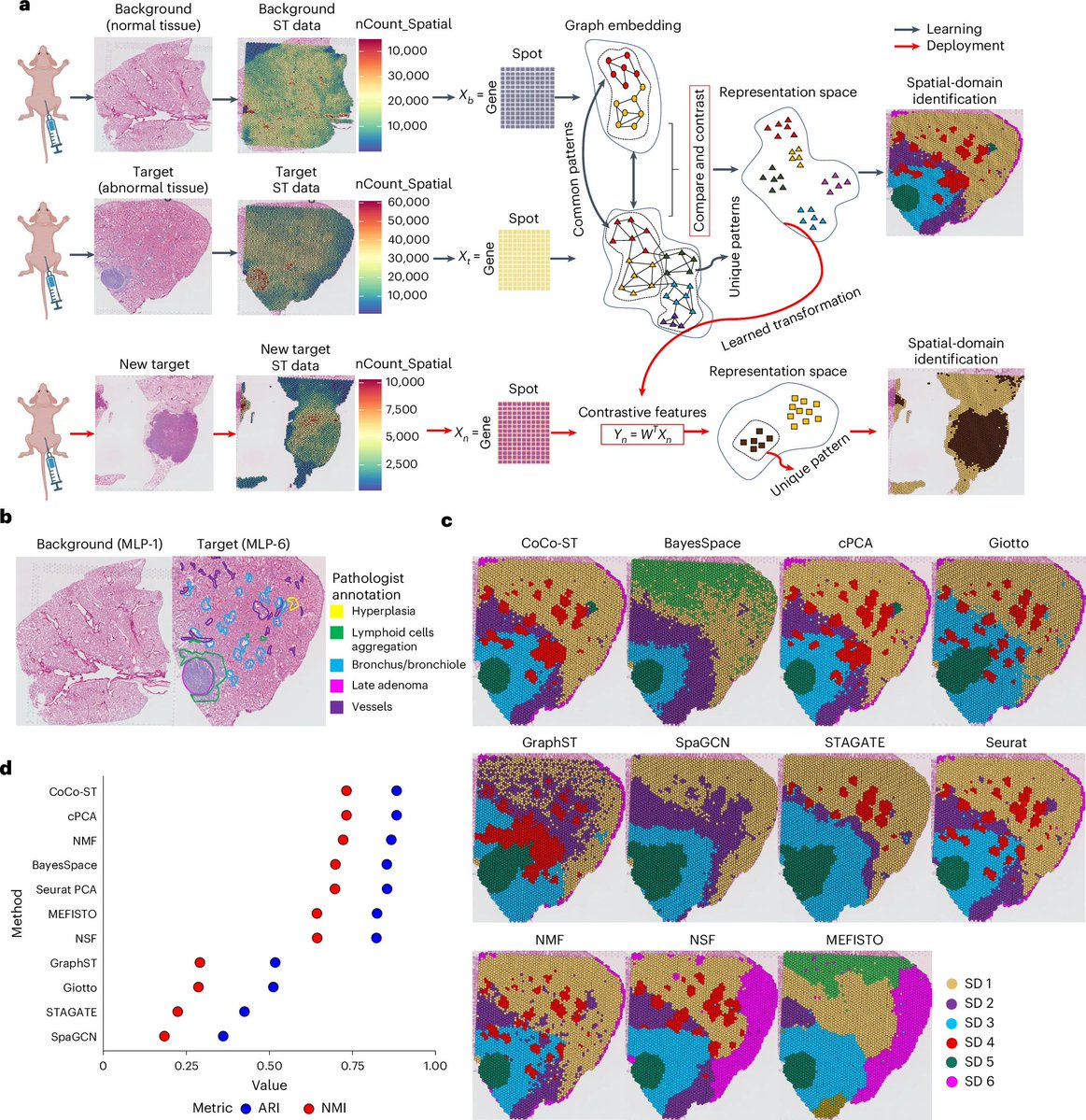
CoCo-ST detects global and local biological structures in spatial transcriptomics datasets. #SpatialTranscriptomics #VarianceIdentification @NatureCellBio nature.com/articles/s4155…
November's #DukeSCIRIP seminar will feature: 1. Xiaohui Jiang of @DukeBiostats on 'X-bridge' for #SingleCell and spatial panel analysis 2. Dr. Ru-Rong Ji of @DukeMedSchool on #SpatialTranscriptomics of #SensoryNeurons 11/5 12-1pm in MSRBIII or online dmpi.duke.edu/duke-single-ce…

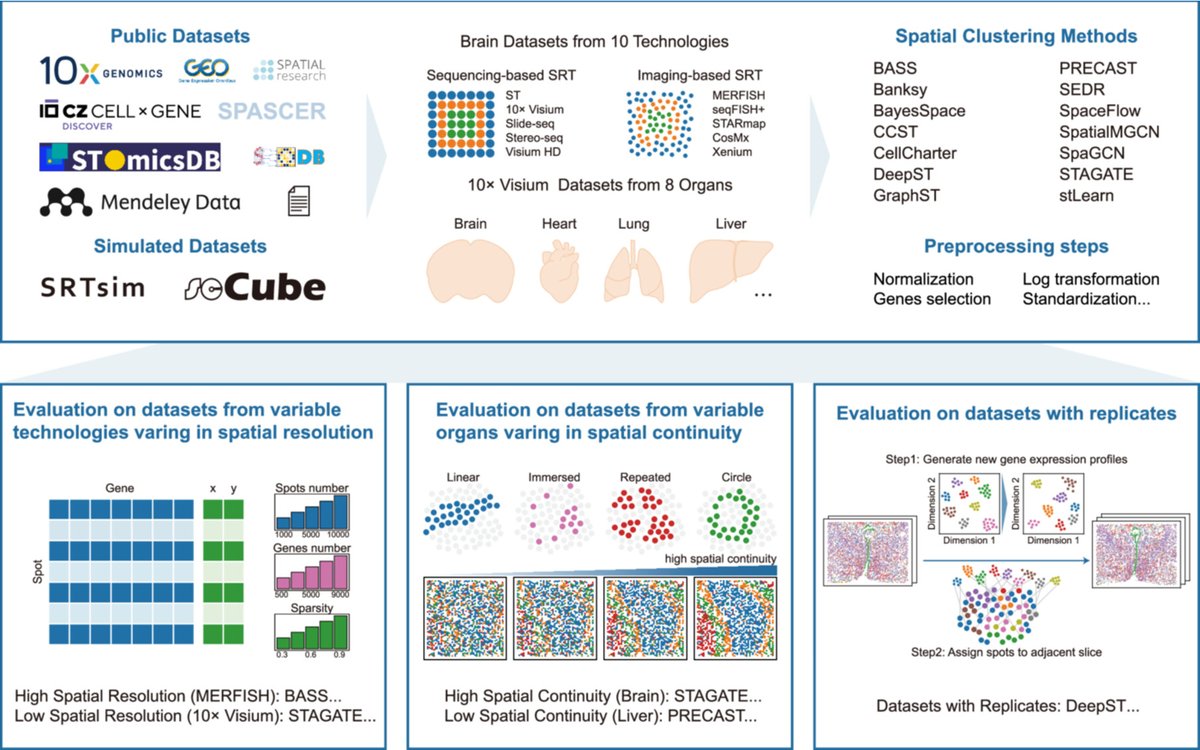
🧬 14 spatial clustering methods benchmarked across ~600 #SpatialTranscriptomics datasets 💻 Covering 10 technologies & 8 organs 📊 Reveals how data traits & spatial patterns shape accuracy ⚙️ Recommends optimal preprocessing & method selection 🔗 doi.org/10.1002/imt2.7……
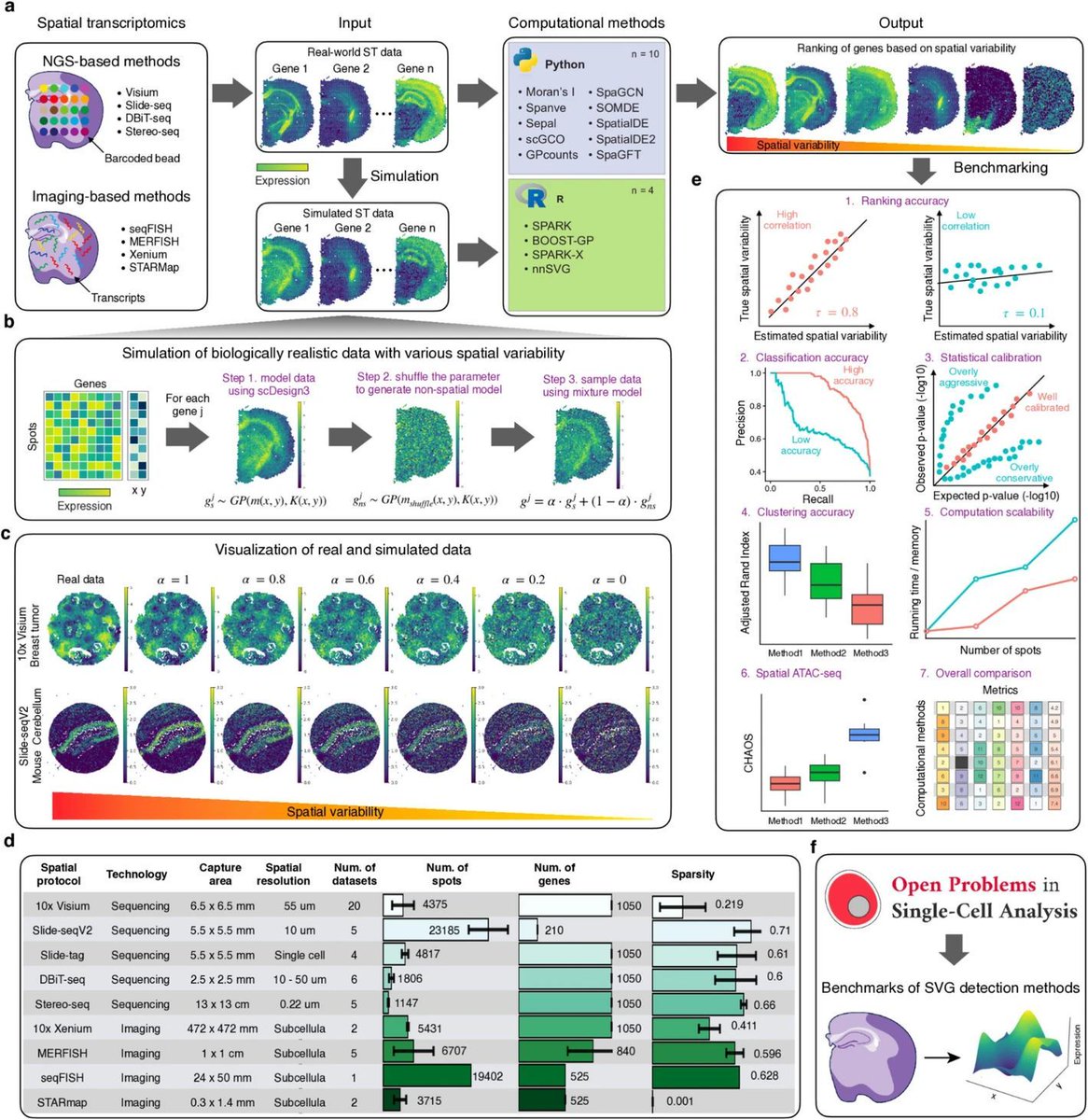
Systematic benchmarking of computational methods to identify spatially variable genes. #SpatialTranscriptomics #MethodsBenchmarking @GenomeBiology genomebiology.biomedcentral.com/articles/10.11…
Accurate cell segmentation is integral to #spatialtranscriptomics—that's why we never stop improving ours. Large cells? Tissues with dense nuclei? We’ve got you covered. See how you can segment cells more accurately than ever before: bit.ly/4m1nEcz


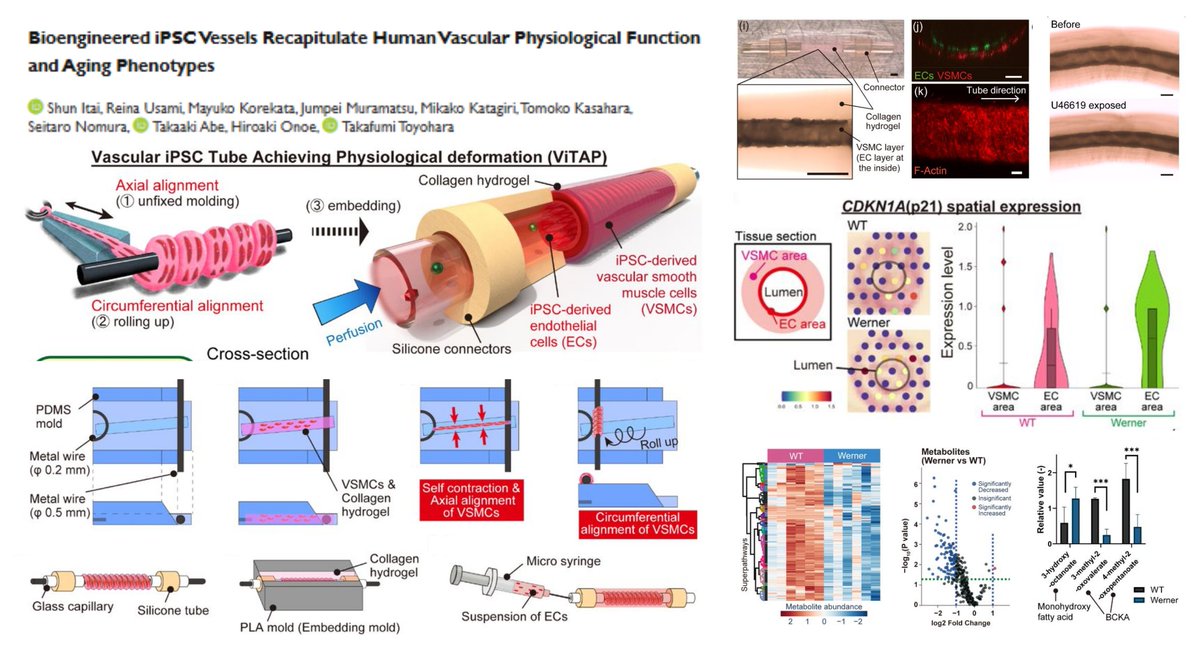
#WernerSyndrome hiPSC-derived engineered vessel #SpatialTranscriptomics #Visium @10xGenomics + #Metabolomics▶️ #BranchedChainKetoAcid⬇️ #MonohydroxyFattyAcid⬆️ in Werner vessels with traits of #PrematureAging Aligned #SmoothMuscleCell in Collagen hydrogel➡️ Roll-up around φ…
⚡️Exciting Breakthrough in Progressive Multiple Sclerosis⚡️ Much humbled to contribute to this exciting project from @Pluchinolab . Analysing 200+ sections from 50+ patients, showcases the power of #spatialtranscriptomics in rare cell localization. 🧠 @IBT_CAS @LukasValihrach
Proud to see our work out in @NeuroCellPress ! cell.com/neuron/fulltex… With Bongsoo Park, Dimitris Tsitsipatis @DanielZucha @LukasValihrach @irina_mohorianu @Pluchinolab @BeermanHSCaging !
JJust released: CosMx Whole Transcriptome Gene List! 🔬 19,000+ genes 🧠 Map pathways, cells & transcripts 📍 Reveal tissue biology at subcellular level 📥 Download now: CosMx Human WTX Gene List | NanoString nanostring.com/resources/cosm… #SpatialTranscriptomics
Comparing single-cell #SpatialTranscriptomics methods on FFPE human tumor #TissueMicroArray CosMx 1k vs MERFISH 500 vs Xenium-UM/-MM 339 93 common genes lung adenocarcinoma pleural mesothelioma Corr. Bulk RNAseq, GeoMx "better F1-scores with Xenium-MM (median, > 75%) than…

How do we uncover cell-cell interactions from #SpatialTranscriptomics data? Excited to share Niche-DE (doi.org/10.1186/s13059…). Niche-DE asks how does a cell’s transcriptome depend on its spatial neighbors, by identifying Niche-Differentially Expressed genes. 1/9

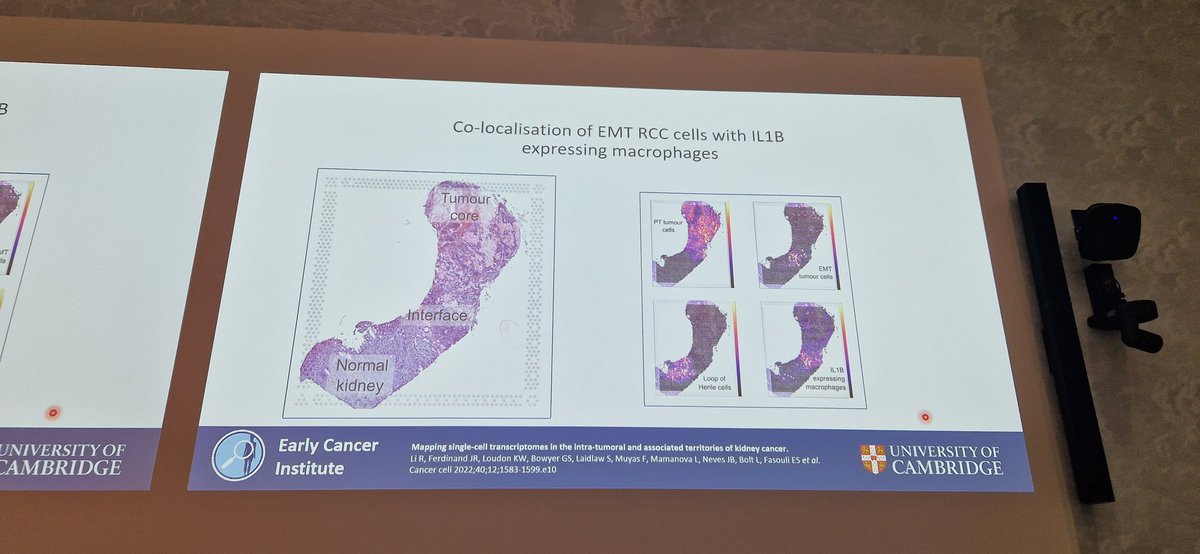
Has @Tom_J_Mitchell finally come round to #SpatialTranscriptomics?! Lovely talk & scope of work presented at #ESUR23 from his @EarlyCancerCam group Using ST to define transition from benign to malignant #KidneyCancer & oppo for early detection/intervention @ChloePacyna @Uroweb


#SpatialTranscriptomics 🐭#Stroke #Visium @10xGenomics #Photothrombosis 3 dpi Stroke▶️ ⏫Microglia/Macrophage Galectin-1/3/9➡️ Astrocyte/Oligodendrocyte CD44 at peri-infarct zone▶️ ⏫remyelination ⏫neurobehavioral performance Post-stroke benefits of LGALS9-expressing…
#SpatialTranscriptomics Visium #CerebralMalaria 🐭+P.berghei ANKA -/+Artesunate Malaria infection▶️ ⏫🧠#EndothelialCell, immune cells SPATA spatial trajectory inference of EC-rich spots in infected🧠▶️ ⏫EC antigen processing/presentation + leukocyte adhesion ⏬BBB Partially…

"We all love the pretty pictures we get from #SpatialTranscriptomics but the real value lies in the ability to precisely determine the cellular composition & cell-cell relationships - not possible with bulk RNA or even scRNA approaches" 💯% well said @bayraktar_lab! 👏 #FoG2025




🚨 Exciting Opportunity! 🚨 @OhioStatePIIO + @OSUWexMed Pathology are recruiting a Tenure-Track Faculty in Spatial Immuno-Oncology Research 🧬🔬 Lead cutting-edge work in #SpatialTranscriptomics, #SpatialProteomics & #ImmunoOncology to decode the #TumorMicroenvironment.…

A spatial long-read approach at near-single-cell resolution reveals developmental regulation of splicing and polyadenylation sites in distinct cortical layers and cell types. #LongReads #SingleCell #Spatialtranscriptomics #IsoSeq #PolyA @NatureComms nature.com/articles/s4146…

Ruochen Dong, @linheng_li lab, gave an outstanding presentation on using #SpatialTranscriptomics to interrogate HSC-niche interaction in the mouse fetal liver at #ISSCR2023. He also received merit and travel awards. Congratulations! @ISSCR

I'm excited to present my first postdoctoral research project: FineST⭐[GitHub:github.com/StatBiomed/Fin…]. Welcome to join and discuss! Your valuable feedback would be greatly appreciated. 🤝 #SpatialTranscriptomics #TumorMicroenvironment #Bioinformatics #CellCommunication
![LingyuLi61's tweet image. I'm excited to present my first postdoctoral research project: FineST⭐[GitHub:github.com/StatBiomed/Fin…]. Welcome to join and discuss! Your valuable feedback would be greatly appreciated. 🤝
#SpatialTranscriptomics #TumorMicroenvironment #Bioinformatics #CellCommunication](https://pbs.twimg.com/media/G36U6HiWsAAt32C.jpg)
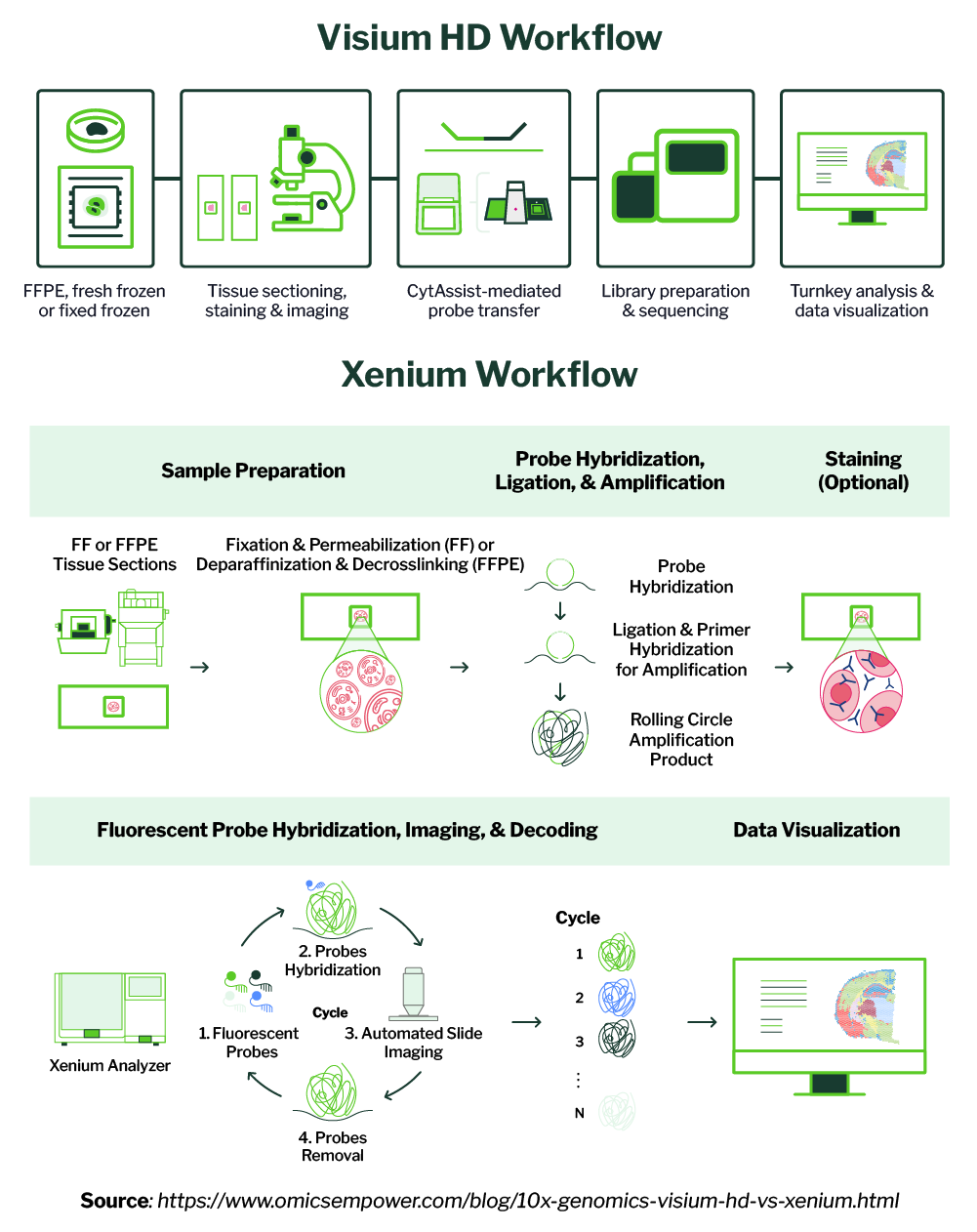
Visium HD or Xenium Prime 5K? Two paths to spatial insights—one for broad, unbiased discovery, the other for targeted, cell-level precision. Read how each fits discovery and validation workflows: signiosbio.com/blog/10x-visiu… #SpatialBiology #SingleCell #SpatialTranscriptomics
Excited to share the fresh STOmics Newsletter (Aug–Sep 2025)! 🎉 220+ Stereo-seq breakthroughs, Alzheimer’s atlas, event recaps & how-to videos—all in one click. Read & retweet! 👉bit.ly/4o0AdXt #STOmics #StereoSeq #SpatialTranscriptomics
🦠💩 #Microbiome 🧬🔬 #SpatialTranscriptomics 🤖🗣️🤖 #ArtificialIntelligence Agents for #PrecisionMedicine & #ColorectalCancer Research 💉 Proud of our #VelazquezVillarrealLab team for presenting TWO outstanding posters on 🎉 2025 @cityofhope Annual Poster Session @PECGSnetwork

November's #DukeSCIRIP seminar will feature: 1. Xiaohui Jiang of @DukeBiostats on 'X-bridge' for #SingleCell and spatial panel analysis 2. Dr. Ru-Rong Ji of @DukeMedSchool on #SpatialTranscriptomics of #SensoryNeurons 11/5 12-1pm in MSRBIII or online dmpi.duke.edu/duke-single-ce…

🧬 14 spatial clustering methods benchmarked across ~600 #SpatialTranscriptomics datasets 💻 Covering 10 technologies & 8 organs 📊 Reveals how data traits & spatial patterns shape accuracy ⚙️ Recommends optimal preprocessing & method selection 🔗 doi.org/10.1002/imt2.7……

CoCo-ST detects global and local biological structures in spatial transcriptomics datasets. #SpatialTranscriptomics #VarianceIdentification @NatureCellBio nature.com/articles/s4155…

Wrapping up an incredible week at #AGTA2025! Grateful to everyone who dropped by our booth. Big thanks to Damian for his talk on the world-first @ParseBio experiment in human malaria–infected spleen samples. #Genomics #SpatialTranscriptomics #EnablingDiscovery




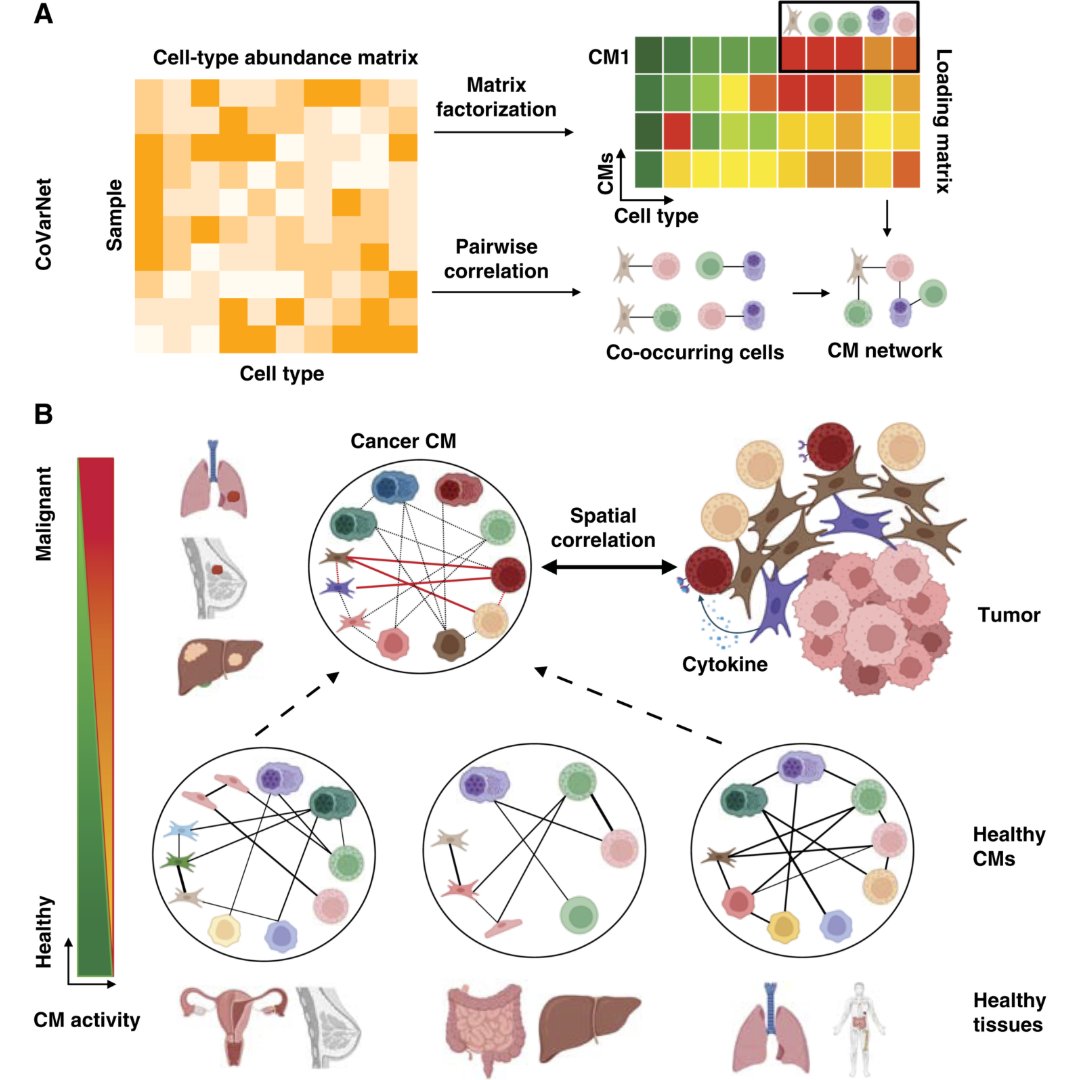
New in the October 15 issue from the Cancer Research special series— From Harmony to Discord: Multicellular Coordination in Tissues and Its Rewiring in Cancer doi.org/10.1158/0008-5… @drmervedede @kchenken @MDAndersonNews #SingleCellTranscriptomics #SpatialTranscriptomics
#SpatialOmics #ImagingMassSpec #SpatialTranscriptomics MISTy Multiview Intercellular SpaTial modeling framework bioconductor.org/packages/relea… View/spatial scale-specific feature interactions Intraview - same location (intra-to-multicellulat) Juxtaview - immediate neighborhood…

⚡️Exciting Breakthrough in Progressive Multiple Sclerosis⚡️ Much humbled to contribute to this exciting project from @Pluchinolab . Analysing 200+ sections from 50+ patients, showcases the power of #spatialtranscriptomics in rare cell localization. 🧠 @IBT_CAS @LukasValihrach
Proud to see our work out in @NeuroCellPress ! cell.com/neuron/fulltex… With Bongsoo Park, Dimitris Tsitsipatis @DanielZucha @LukasValihrach @irina_mohorianu @Pluchinolab @BeermanHSCaging !
ChatGPT-Like AI Model Details 1,300 Regions/Subregions in Mouse Brain Map Tissue-agnostic CellTransformer can be used on other organs and tissues, including cancerous tissue, for which large-scale #spatialtranscriptomics data is available @UCSF #brain #AI hubs.li/Q03MQQXq0
🚀 Pilot Now Open: @ST_Omics OMNI with @SA_genomics & Decode Science Perfect for generating pilot data, exploring new projects, and strengthening grant applications. 🔗 Learn more: app-ap1.hubspotdocuments.com/documents/2389… #SpatialTranscriptomics #SingleCell #Genomics #EnablingDiscovery
Now Open! STOmics OMNI pilot - SAGC, @Decode_Science - spatial RNA from FFPE—end-to-end service. Why STOmics OMNI? ✅ Species-agnostic ✅ Random hexamers (total RNA) ✅ 10×10 mm area at high res (500 nm) ✅ End-to-end workflow Get your projects ready→ap1.hubs.ly/y0hZyD0

#WernerSyndrome hiPSC-derived engineered vessel #SpatialTranscriptomics #Visium @10xGenomics + #Metabolomics▶️ #BranchedChainKetoAcid⬇️ #MonohydroxyFattyAcid⬆️ in Werner vessels with traits of #PrematureAging Aligned #SmoothMuscleCell in Collagen hydrogel➡️ Roll-up around φ…

Accurate cell segmentation is integral to #spatialtranscriptomics—that's why we never stop improving ours. Large cells? Tissues with dense nuclei? We’ve got you covered. See how you can segment cells more accurately than ever before: bit.ly/4m1nEcz


🚨New #SpatialTranscriptomics #Bioinformatics data resource out in @naturemethods. SODB, a platform with >2,400 manually curated spatial experiments from >25 spatial omics technologies & interactive analytical modules. This🧵will walk you through all the features of SODB [1/33]
![simocristea's tweet image. 🚨New #SpatialTranscriptomics #Bioinformatics data resource out in @naturemethods.
SODB, a platform with >2,400 manually curated spatial experiments from >25 spatial omics technologies & interactive analytical modules.
This🧵will walk you through all the features of SODB [1/33]](https://pbs.twimg.com/media/FpqfK6HaUAAQZAo.jpg)
Ruochen Dong, @linheng_li lab, gave an outstanding presentation on using #SpatialTranscriptomics to interrogate HSC-niche interaction in the mouse fetal liver at #ISSCR2023. He also received merit and travel awards. Congratulations! @ISSCR

CoCo-ST detects global and local biological structures in spatial transcriptomics datasets. #SpatialTranscriptomics #VarianceIdentification @NatureCellBio nature.com/articles/s4155…

Wow, this is big! Congrats to all authors!👏 👉 Spatial and single-nucleus transcriptomic analysis of genetic and sporadic forms of Alzheimer's Disease 👉 biorxiv.org/content/10.110… #SpatialTranscriptomics #Visium #snRNAseq #ParseBiosciences

Our work on spatial multiomics in Alzheimer’s disease and Down syndrome now on bioRxiv biorxiv.org/content/10.110…
How do we uncover cell-cell interactions from #SpatialTranscriptomics data? Excited to share Niche-DE (doi.org/10.1186/s13059…). Niche-DE asks how does a cell’s transcriptome depend on its spatial neighbors, by identifying Niche-Differentially Expressed genes. 1/9

🧬 14 spatial clustering methods benchmarked across ~600 #SpatialTranscriptomics datasets 💻 Covering 10 technologies & 8 organs 📊 Reveals how data traits & spatial patterns shape accuracy ⚙️ Recommends optimal preprocessing & method selection 🔗 doi.org/10.1002/imt2.7……

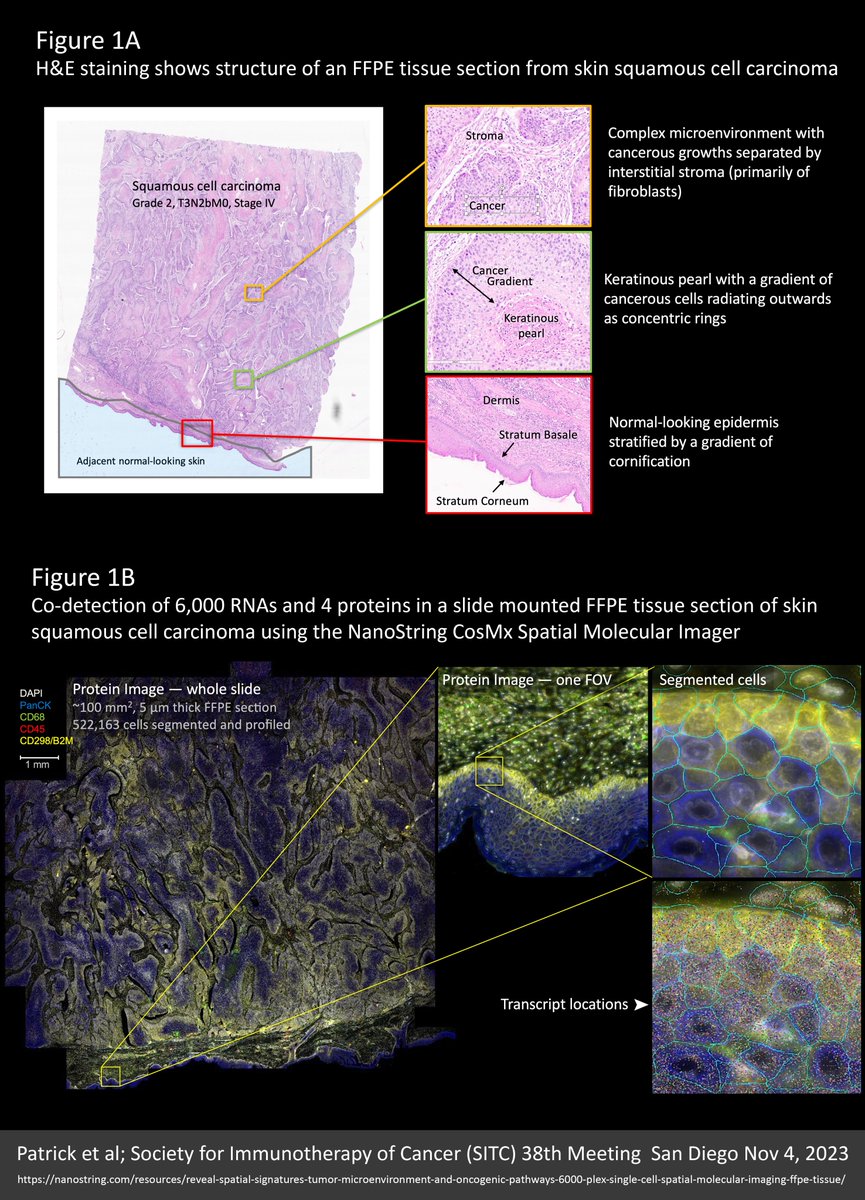
We’re all wondering what the future of #spatialtranscriptomics (ST) has in store. Well, I think the near future will look a lot like the new 6000-plex dataset reviewed here (link to the original source at the end of this post). SAMPLE An entire 100 mm² intact 5 µm FFPE section…



#SpatialTranscriptomics 🐭#Stroke #Visium @10xGenomics #Photothrombosis 3 dpi Stroke▶️ ⏫Microglia/Macrophage Galectin-1/3/9➡️ Astrocyte/Oligodendrocyte CD44 at peri-infarct zone▶️ ⏫remyelination ⏫neurobehavioral performance Post-stroke benefits of LGALS9-expressing…

#WernerSyndrome hiPSC-derived engineered vessel #SpatialTranscriptomics #Visium @10xGenomics + #Metabolomics▶️ #BranchedChainKetoAcid⬇️ #MonohydroxyFattyAcid⬆️ in Werner vessels with traits of #PrematureAging Aligned #SmoothMuscleCell in Collagen hydrogel➡️ Roll-up around φ…

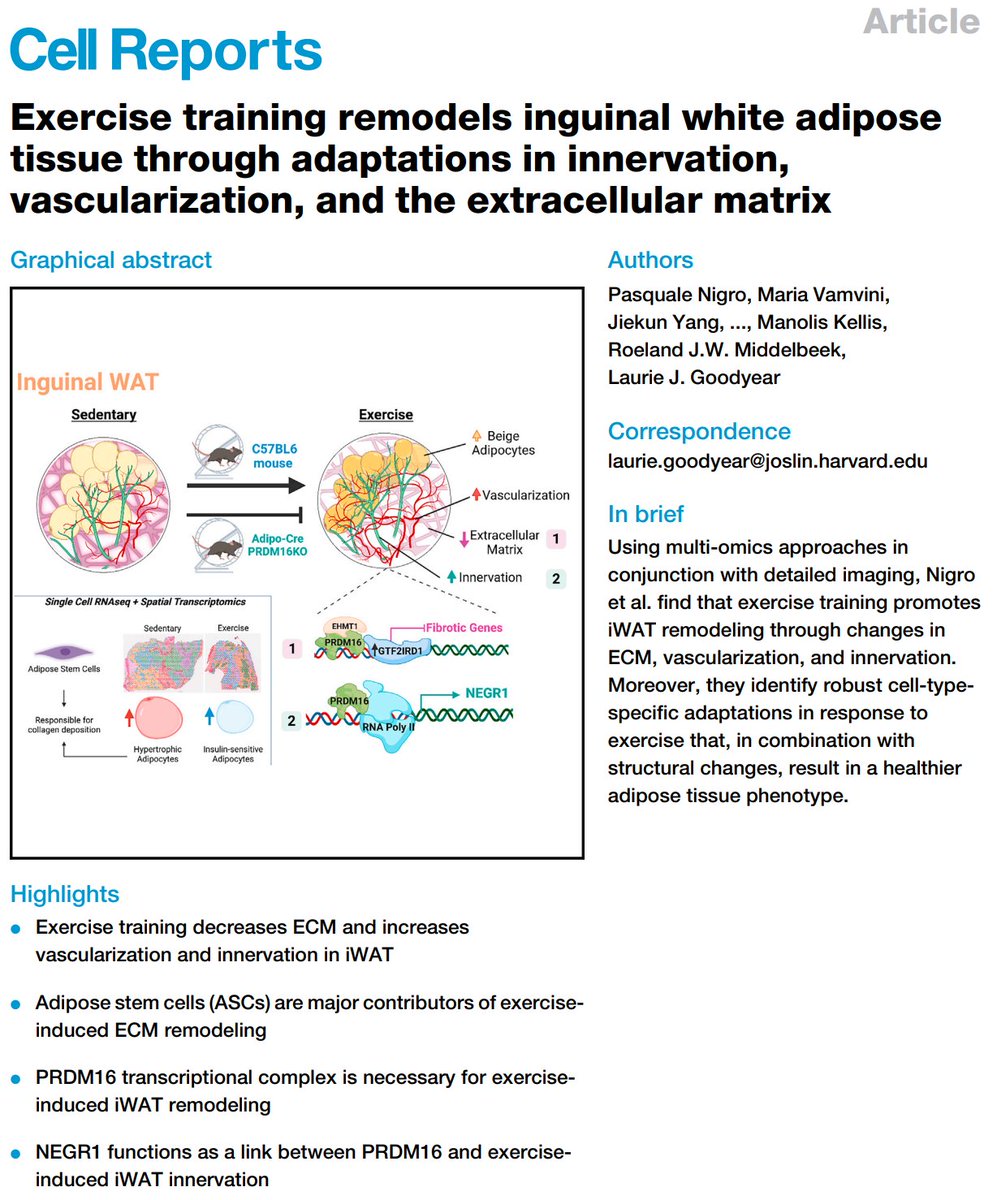
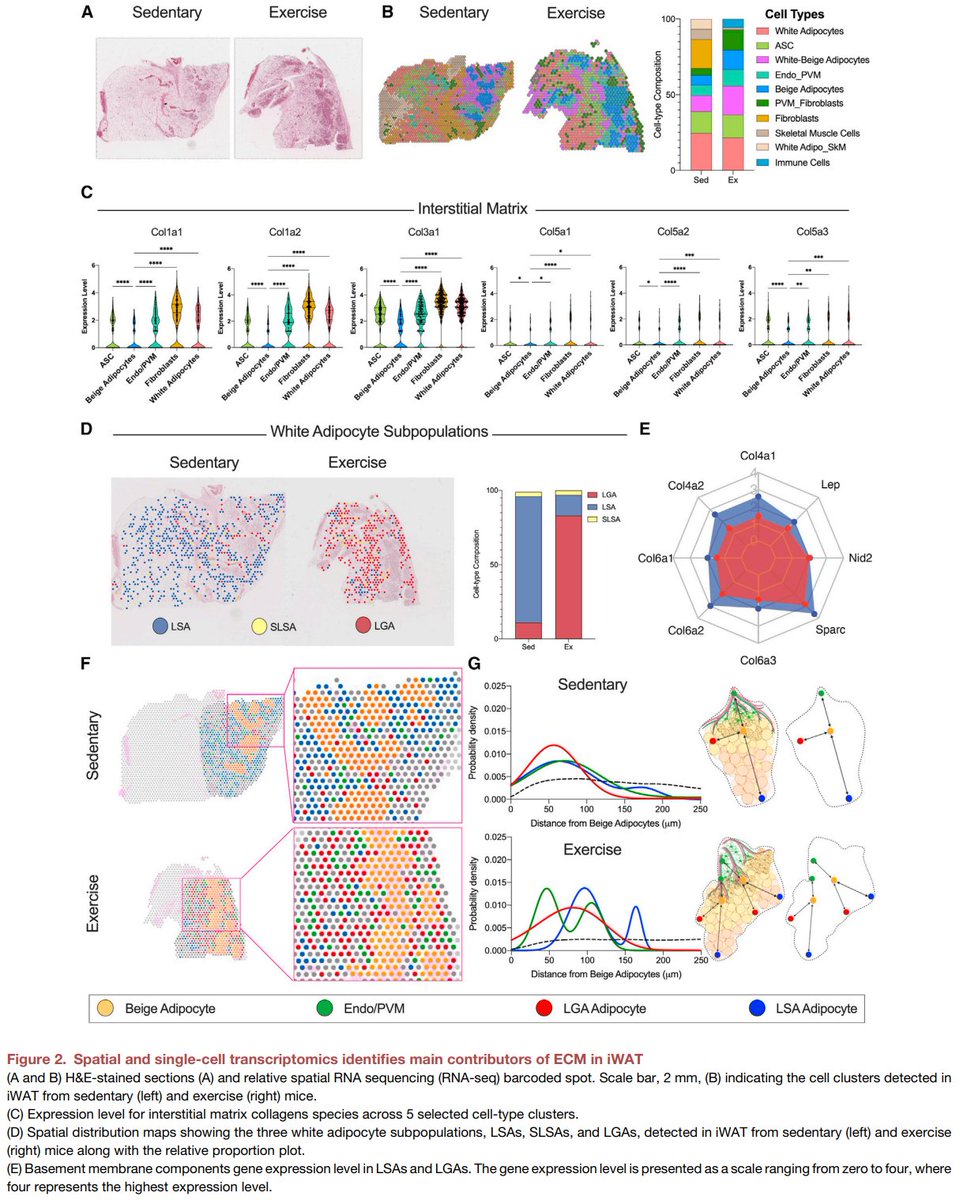
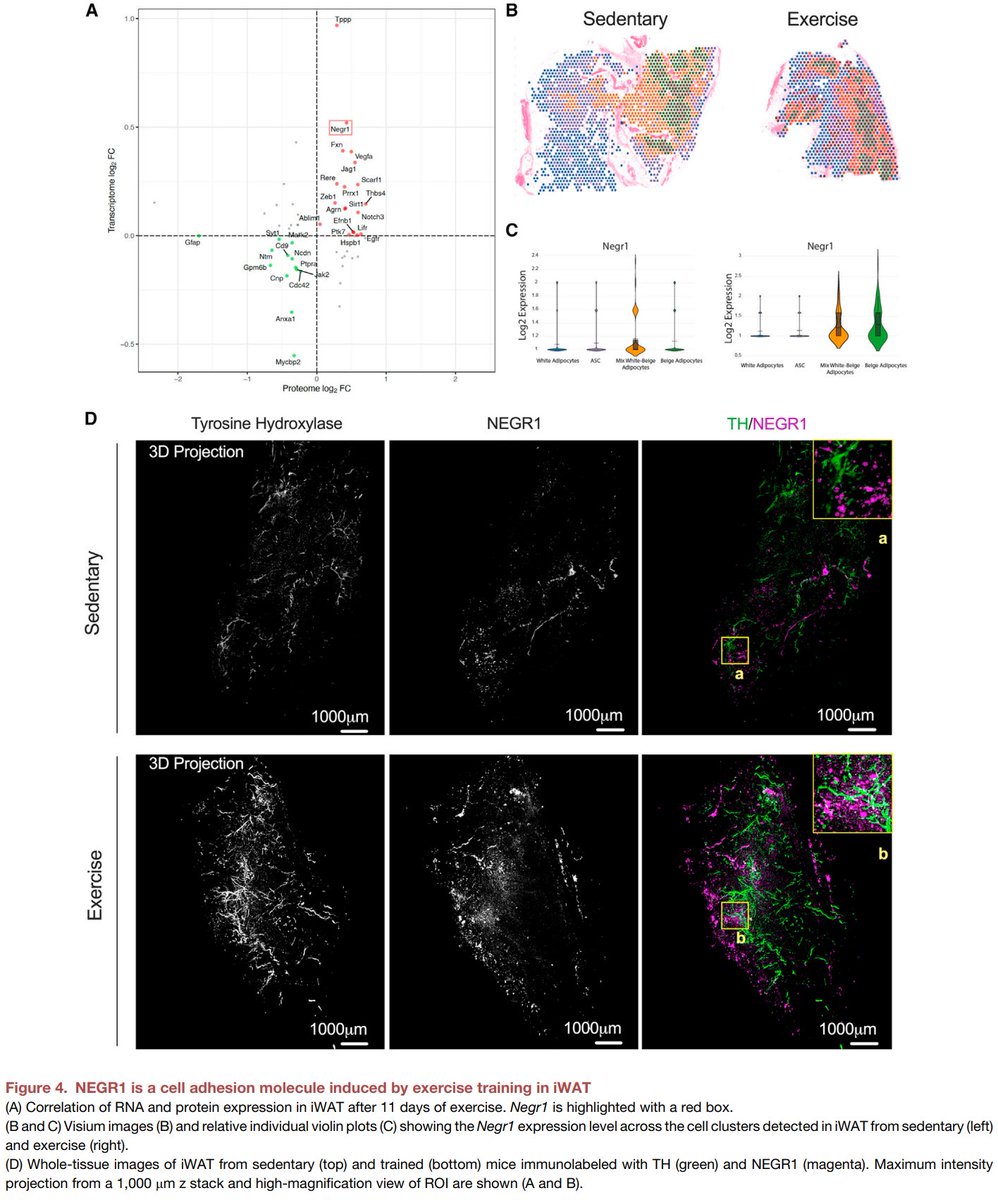
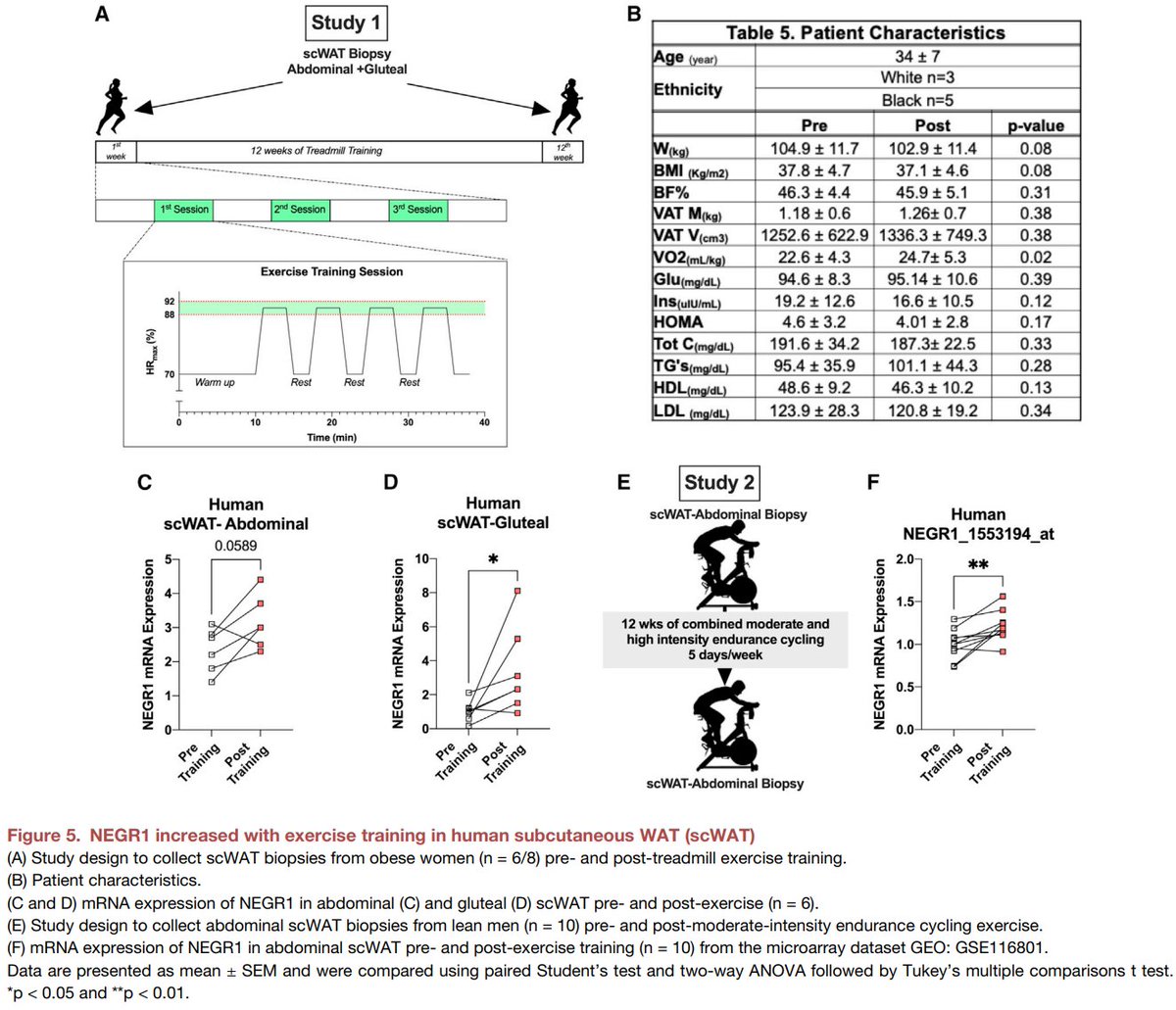
Excited to share our #SingleCell and #SpatialTranscriptomics analysis of #ExerciseTraining remodeling of #WhiteAdiposeTissue through innervation, vascularization, and #ExtracellularMatrix in mouse and human with #LaurieGoodyear #JanWillemMiddelbeek @PasqualeNigro9 @MVamvini…
Comparing single-cell #SpatialTranscriptomics methods on FFPE human tumor #TissueMicroArray CosMx 1k vs MERFISH 500 vs Xenium-UM/-MM 339 93 common genes lung adenocarcinoma pleural mesothelioma Corr. Bulk RNAseq, GeoMx "better F1-scores with Xenium-MM (median, > 75%) than…

A spatial long-read approach at near-single-cell resolution reveals developmental regulation of splicing and polyadenylation sites in distinct cortical layers and cell types. #LongReads #SingleCell #Spatialtranscriptomics #IsoSeq #PolyA @NatureComms nature.com/articles/s4146…

High-resolution mapping of single cells in spatial context. #SingleCell #SpatialTranscriptomics #Bioinformatics @NatureComms nature.com/articles/s4146…
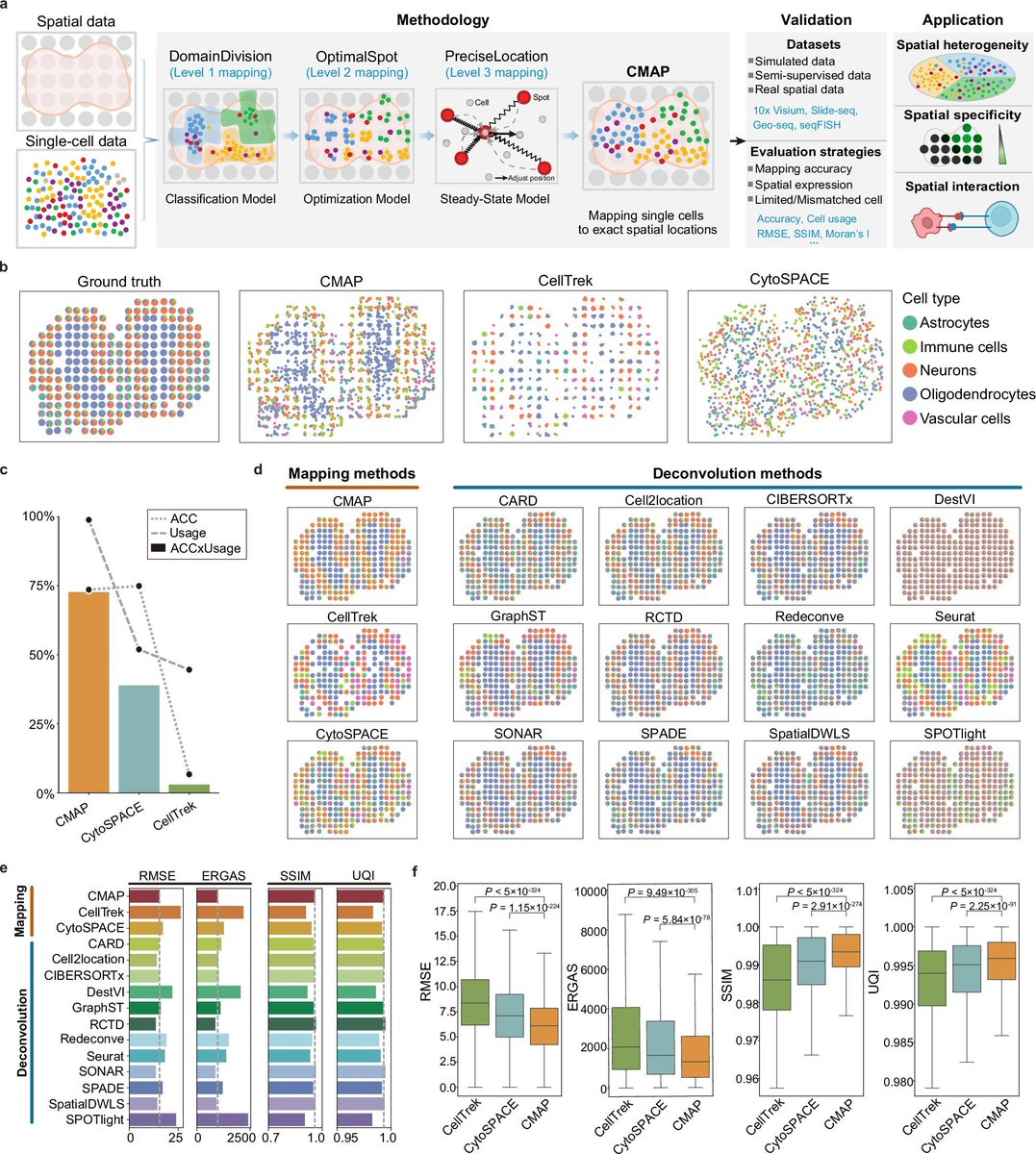
Now online in @CD_AACR: #SpatialTranscriptomics of Intraductal Papillary Mucinous Neoplasms (#IPMN) of the Pancreas Identifies NKX6-2 as a Driver of Gastric Differentiation and Indolent Biological Potential @Marta_Sans_ @Aiims1742 @MDAndersonNews doi.org/10.1158/2159-8…

We are very very VERY excited to be running #cosmx #singlecell #spatialtranscriptomics experiments in @UofGlasgow @UofGCancerSci @UofGMVLS Well done to the @LabSpatialNBJ team @nanostringtech


Excellent talk from @Sandy_Figiel in @Uroweb #EAUlab session at #EAU25 presenting our cool data (I'm bias!) on #SpatialTranscriptomics & #ClonalAnalysis to detect unique features of metastasing clones in #ProstateCancer. @OxPCaBiol #SPACE_Study #3DLightsheet




🔬 A new computational approach pioneered by researchers at the @broadinstitute eliminates time-intensive imaging, enabling high-resolution spatial mapping of gene expression in tissue. See how they're making #SpatialTranscriptomics easier for everyone: bit.ly/3G9Hrau

#SpatialTranscriptomics Visium #CerebralMalaria 🐭+P.berghei ANKA -/+Artesunate Malaria infection▶️ ⏫🧠#EndothelialCell, immune cells SPATA spatial trajectory inference of EC-rich spots in infected🧠▶️ ⏫EC antigen processing/presentation + leukocyte adhesion ⏬BBB Partially…

Something went wrong.
Something went wrong.
United States Trends
- 1. Aaron Gordon 25.4K posts
- 2. Steph 60.5K posts
- 3. Jokic 22.8K posts
- 4. #criticalrolespoilers 12.3K posts
- 5. Halle 19.8K posts
- 6. Wentz 25.2K posts
- 7. Vikings 52.6K posts
- 8. Warriors 86.6K posts
- 9. #EAT_IT_UP_SPAGHETTI 228K posts
- 10. #DubNation 4,453 posts
- 11. Hobi 40.2K posts
- 12. #LOVERGIRL 17.6K posts
- 13. Nuggets 26K posts
- 14. Cam Johnson 1,634 posts
- 15. Chargers 57.7K posts
- 16. Megan 37.2K posts
- 17. Pacers 22.4K posts
- 18. Shai 25.4K posts
- 19. Kuminga 5,647 posts
- 20. Brosmer 3,915 posts