#spatialtranscriptomics 搜尋結果
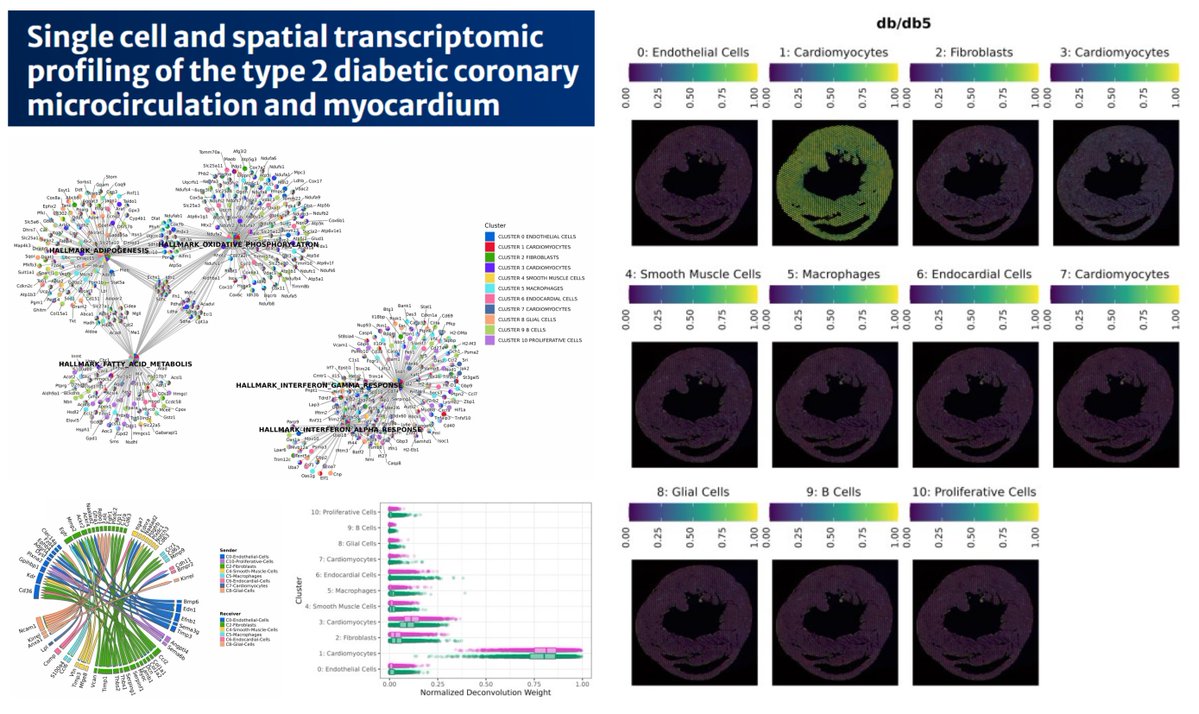
Diabetic #T2D db/db 🐭 heart-coronary resistance microvessel multiomics FFPE-scRNAseq #SpatialTranscriptomics Visium @10xGenomics Proteomics Septal Coronary resistance microvessels (how to isolate?😮) Traits of Fatty acid β-oxidation & ketogenesis (Hmgcs2, Acot2) in diabetic…
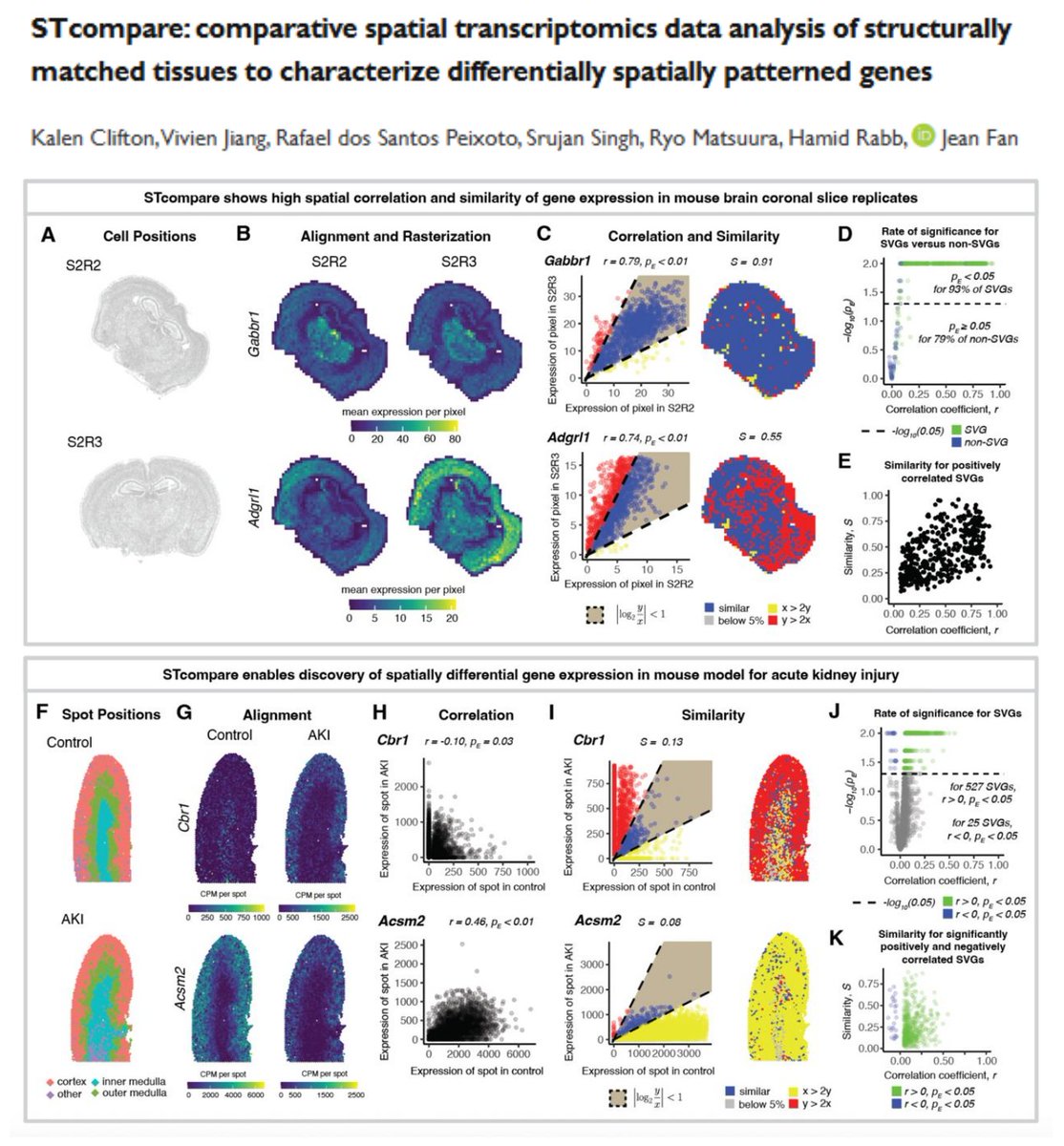
#SpatialTranscriptomics STcompare github.com/JEFworks-Lab/S… Test your gene of interest if it is differentially spatially patterned (r, p, s)🤠 Spatial correlation Spatial fold change Control for spatial autocorrelation Comparisons will be straightforward with Alignment (STalign)…
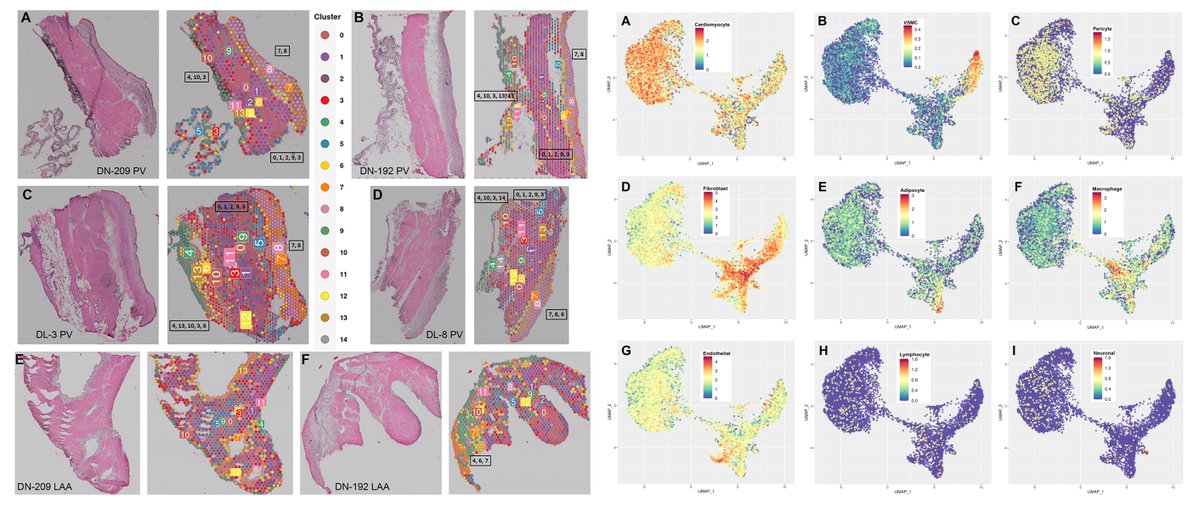
#SpatialTranscriptomics Visium @10xGenomics Human Left Atrial Appendage (n=2 donors) & Pulmonary Vein (n=8 donors) Spatial location of #AtrialFibrillation risk genes PITX2 SHOX2 HCN4 @CVpathol 2025 sciencedirect.com/science/articl…
🧬 IICD Monthly Research Roundup – November 2025 #IICDResearch #CancerGenomics #SpatialTranscriptomics #CancerResearch


New publication by our student, Odmaa Bayaraa: 'Multi-region #SpatialTranscriptomics reveals region specific differences in response to amyloid beta (Aβ) plaque induced changes in #Alzheimers disease (#AD)' So proud of this work & Odmaa! Check it out 🧬🧠 pubmed.ncbi.nlm.nih.gov/41318546/
Accurately deciphering the spatial gene expression characteristics of individual cells represents a core demand in spatial transcriptomics research. #SpatialTranscriptomics #CellSegmentationTech #SpatialOmics #SingleCellSpatialExpression

Unlocking the true power of precision therapeutics: Spatial RNA medicine maps gene expression within tissues to target treatments exactly where they're needed. 🧬🔬 Curious how this changes drug design? Read more: news-medical.net/life-sciences/… #PrecisionMedicine #SpatialTranscriptomics
Something went wrong.
Something went wrong.
United States Trends
- 1. FINALLY DID IT 622K posts
- 2. The JUP 180K posts
- 3. The PENGU 227K posts
- 4. The BONK 170K posts
- 5. Good Tuesday 32.6K posts
- 6. #PersonOfTheYearAwards2025 475K posts
- 7. Alex Delgado N/A
- 8. #riddle 3,715 posts
- 9. #amulet 3,207 posts
- 10. #MAGMA N/A
- 11. #jovian 3,443 posts
- 12. Namjoon 86.7K posts
- 13. DataHaven 26K posts
- 14. OT7 LIVE 41.8K posts
- 15. Rodman 1,623 posts
- 16. TOP CALL 8,602 posts
- 17. Jokic 11.9K posts
- 18. The BBC 117K posts
- 19. Rockets 16.9K posts
- 20. Sengun 6,230 posts