#spatialtranscriptomics результаты поиска
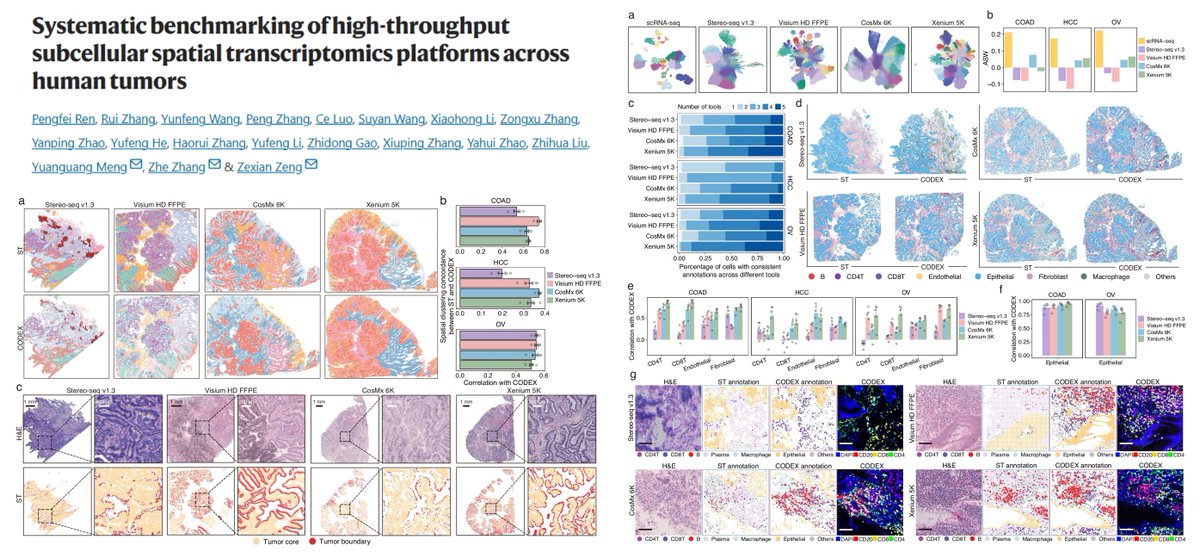
Systematic comparisons among 4 subcellular-resolution #SpatialTranscriptomics methods Stereo-seq v1.3 Visium HD FFPE CosMx 6k Xenium 5k Human Tumors Ground truth CODEX, +scRNAseq Gene detection sensitivity (CosMx performance...🙁) (Spatial) false positives Transcript-protein…
🚨New #SpatialTranscriptomics #Bioinformatics data resource out in @naturemethods. SODB, a platform with >2,400 manually curated spatial experiments from >25 spatial omics technologies & interactive analytical modules. This🧵will walk you through all the features of SODB [1/33]
![simocristea's tweet image. 🚨New #SpatialTranscriptomics #Bioinformatics data resource out in @naturemethods.
SODB, a platform with >2,400 manually curated spatial experiments from >25 spatial omics technologies & interactive analytical modules.
This🧵will walk you through all the features of SODB [1/33]](https://pbs.twimg.com/media/FpqfK6HaUAAQZAo.jpg)
Ruochen Dong, @linheng_li lab, gave an outstanding presentation on using #SpatialTranscriptomics to interrogate HSC-niche interaction in the mouse fetal liver at #ISSCR2023. He also received merit and travel awards. Congratulations! @ISSCR

Comparing single-cell #SpatialTranscriptomics methods on FFPE human tumor #TissueMicroArray CosMx 1k vs MERFISH 500 vs Xenium-UM/-MM 339 93 common genes lung adenocarcinoma pleural mesothelioma Corr. Bulk RNAseq, GeoMx "better F1-scores with Xenium-MM (median, > 75%) than…

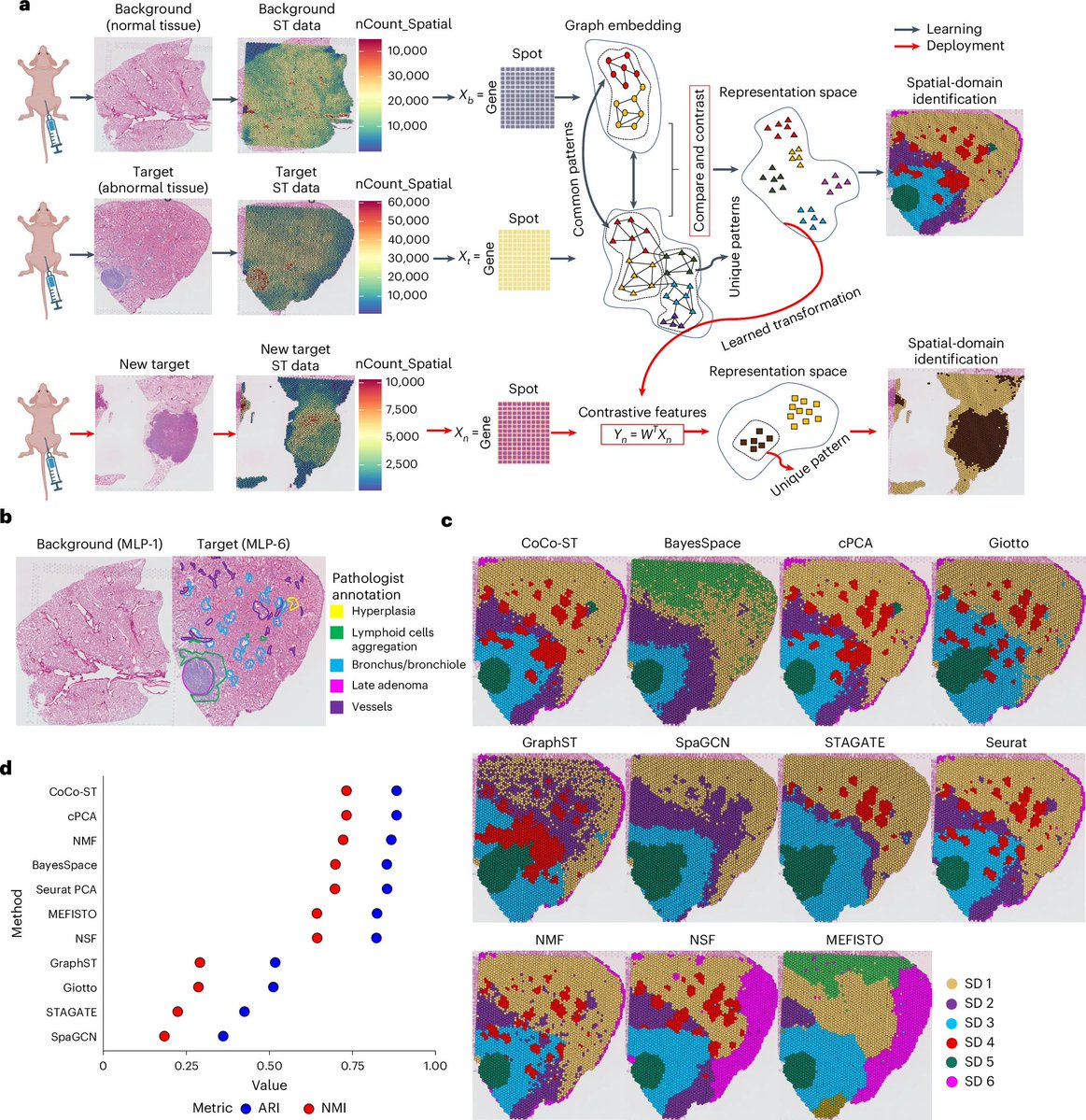
CoCo-ST detects global and local biological structures in spatial transcriptomics datasets. #SpatialTranscriptomics #VarianceIdentification @NatureCellBio nature.com/articles/s4155…
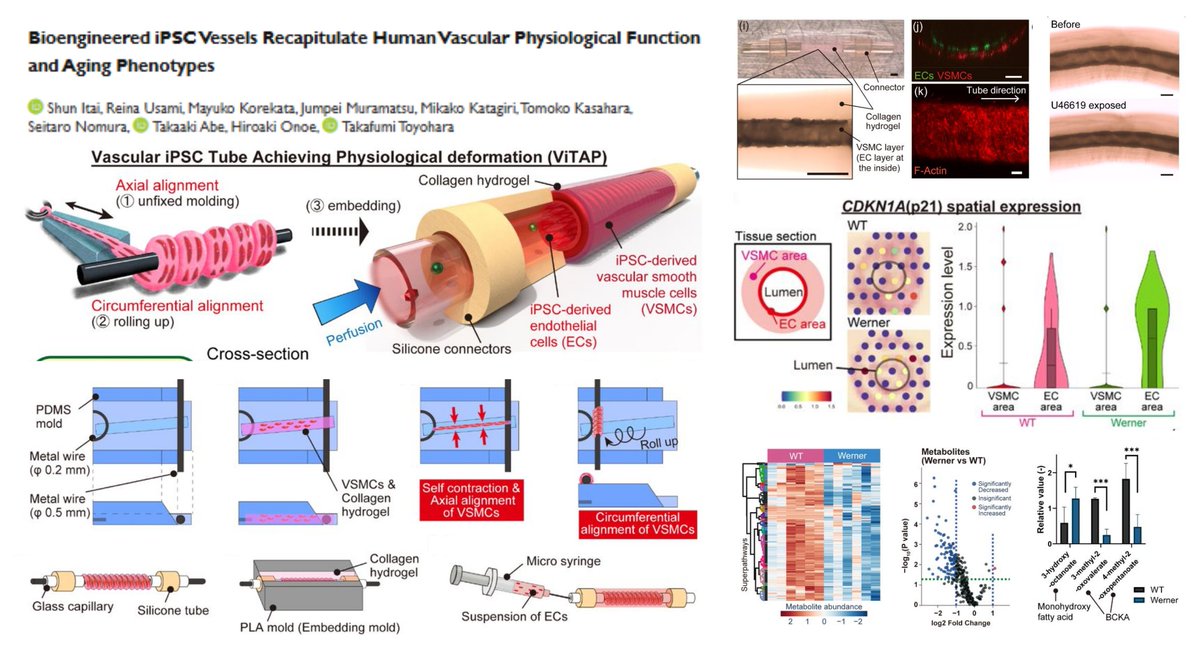
#WernerSyndrome hiPSC-derived engineered vessel #SpatialTranscriptomics #Visium @10xGenomics + #Metabolomics▶️ #BranchedChainKetoAcid⬇️ #MonohydroxyFattyAcid⬆️ in Werner vessels with traits of #PrematureAging Aligned #SmoothMuscleCell in Collagen hydrogel➡️ Roll-up around φ…
We are very very VERY excited to be running #cosmx #singlecell #spatialtranscriptomics experiments in @UofGlasgow @UofGCancerSci @UofGMVLS Well done to the @LabSpatialNBJ team @nanostringtech


🔬 A new computational approach pioneered by researchers at the @broadinstitute eliminates time-intensive imaging, enabling high-resolution spatial mapping of gene expression in tissue. See how they're making #SpatialTranscriptomics easier for everyone: bit.ly/3G9Hrau

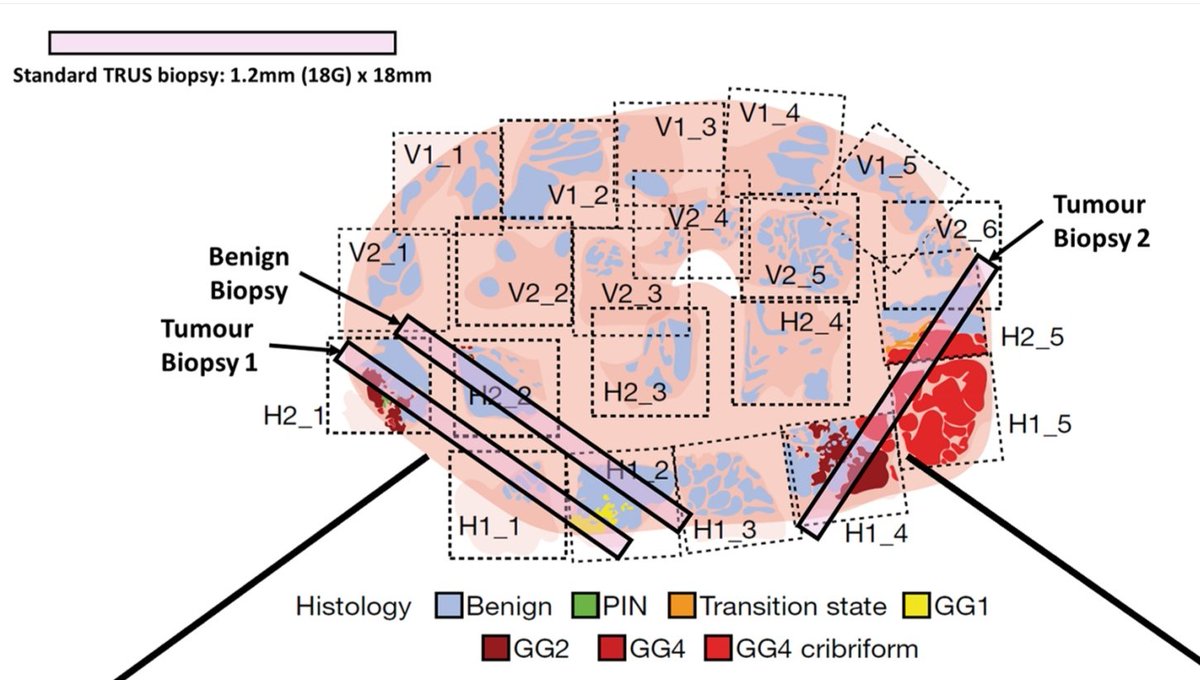
Excellent talk from @Sandy_Figiel in @Uroweb #EAUlab session at #EAU25 presenting our cool data (I'm bias!) on #SpatialTranscriptomics & #ClonalAnalysis to detect unique features of metastasing clones in #ProstateCancer. @OxPCaBiol #SPACE_Study #3DLightsheet




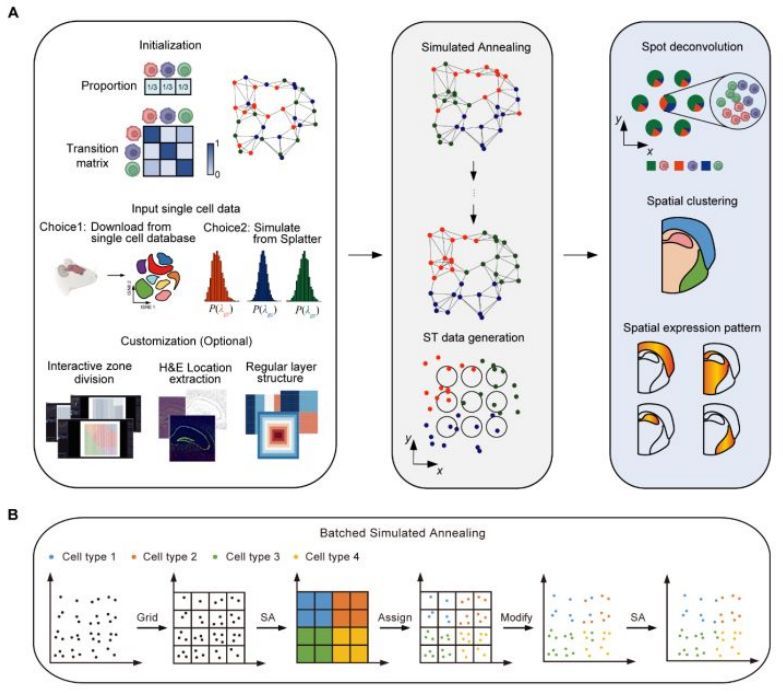
Spider: a flexible and unified framework for simulating spatial transcriptomics data. #SpatialTranscriptomics #DataSimulation #Genomics #Bioinformatics 🧬 🖥️ academic.oup.com/bioinformatics…
#SpatialTranscriptomics #ParkinsonsDisease A role of #FGF9 in Glia #Ferroptosis in PD?😁 #Visium @10xGenomics +snRNAseq MPTP🐭 #SubstantiaNigra FGF9 ⏬in PD Neuron ⏫in PD Ependymal cell FGF9-to-FGFR1/2/3 (CellChat, likely Neuron-to-Oligodendrocyte/Astrocyte) ⏬in PD…

Vispro improves imaging analysis for Visium spatial transcriptomics.#SpatialTranscriptomics #Visium #ImageProcessing #Bioinformatics #Genomics @GenomeBiology genomebiology.biomedcentral.com/articles/10.11…
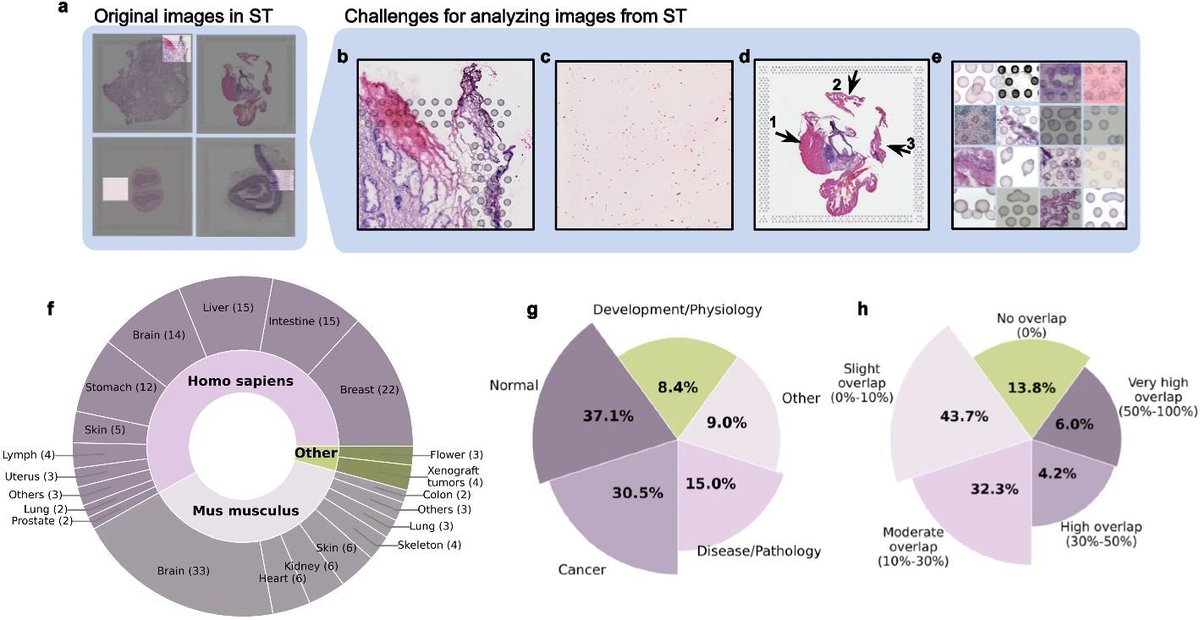
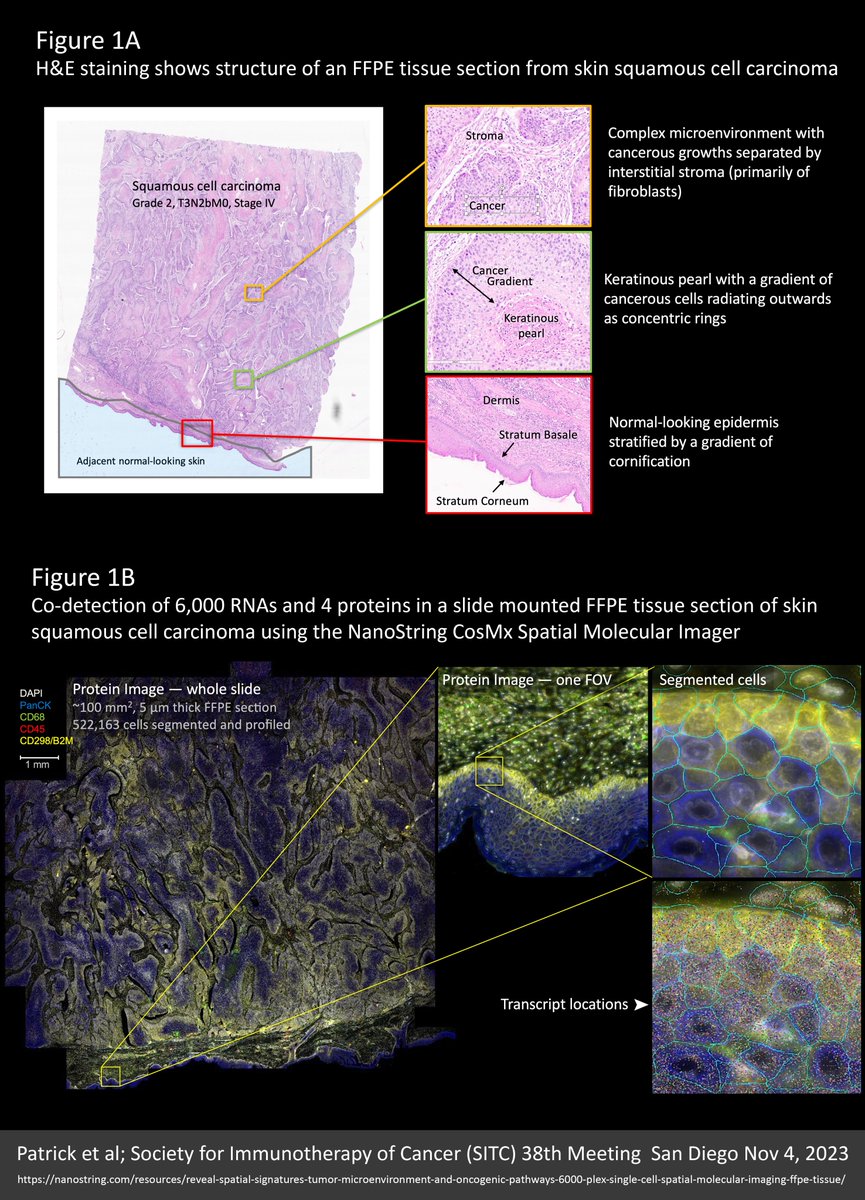
We’re all wondering what the future of #spatialtranscriptomics (ST) has in store. Well, I think the near future will look a lot like the new 6000-plex dataset reviewed here (link to the original source at the end of this post). SAMPLE An entire 100 mm² intact 5 µm FFPE section…



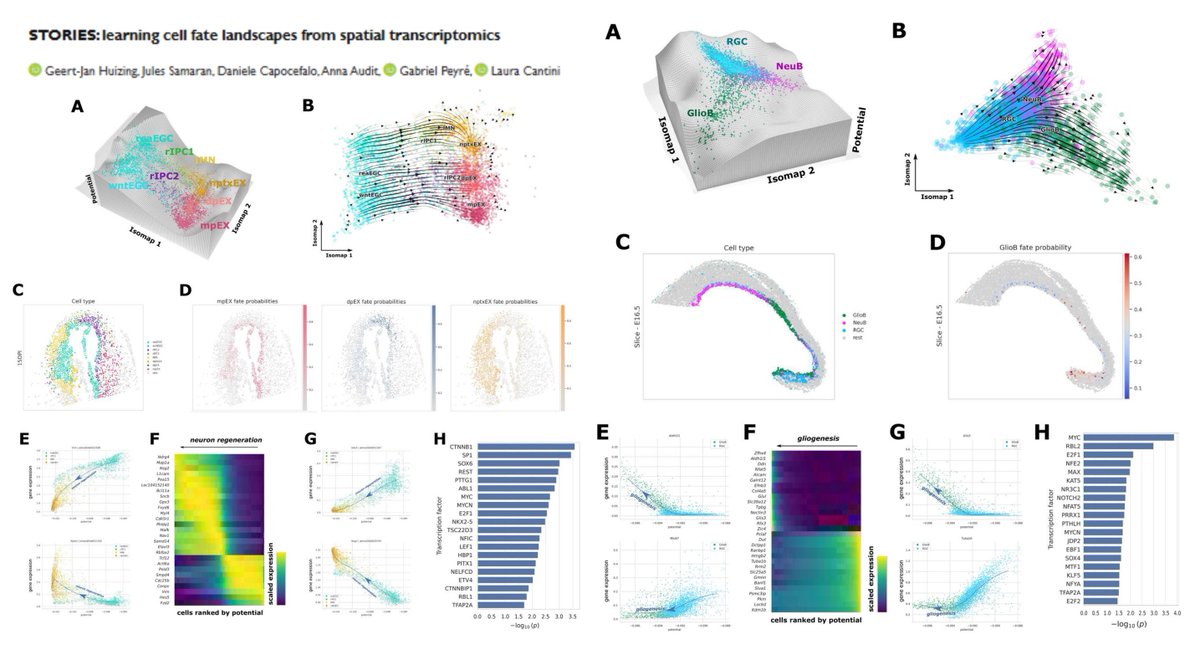
STORIES SpatioTemporal Omics eneRgIES Learning Waddington epigenetic (+regulon) landscape from multi-time-point Stereo-seq #SpatialTranscriptomics Potential-based Fused Gromov-Wasserstein #OptimalTransport Linear▶️gene expression error Quadratic▶️spatial consistency @gjhuizing…
Pleased to share our latest #SpatialTranscriptomics output now on @biorxivpreprint We show marked variation in gene expr relating to @Decipher_VCYT, #Oncotype_DX & #ProstaDiag genomic scores Being presented by @Sandy_Figiel this afternoon #EAU23!! biorxiv.org/content/10.110…



In our latest study, we look at how storing kidneys on ice before transplant can cause damage over time. We use #spatialtranscriptomics to characterize cold ischemia injury at multiple time points to reveal compartment-specific changes Preprint: biorxiv.org/content/10.110… 🧵👇 1/n

"We all love the pretty pictures we get from #SpatialTranscriptomics but the real value lies in the ability to precisely determine the cellular composition & cell-cell relationships - not possible with bulk RNA or even scRNA approaches" 💯% well said @bayraktar_lab! 👏 #FoG2025




1st single-cell #SpatialTranscriptomics map of human coronary #Atherosclerosis published in peer-reviewed format🤠 CosMx 1k + GeoMx @brukerspatial Immune cell-focused analysis Use CosMx data to deconvolve GeoMx cell composition...😆 @EmboMolMed 2025 embopress.org/doi/full/10.10…

Congrats to @Sandy_Figiel for winning poster 1st Prize (out of 53 posters!) at #OxSymp23 @OxfordCancer. Using #SpatialTranscriptomics to reveal problem of heterogeneity when using genomic prognostic scores in #ProstateCancer #Organscale Full preprint: biorxiv.org/content/10.110…




Something went wrong.
Something went wrong.
United States Trends
- 1. The BONK 83.4K posts
- 2. Good Thursday 32.5K posts
- 3. #thursdayvibes 1,792 posts
- 4. Happy Friday Eve N/A
- 5. Usher 4,239 posts
- 6. Shaggy 2,454 posts
- 7. Godzilla 26K posts
- 8. #PiratasDelImperio 1,601 posts
- 9. #ThursdayThoughts 2,124 posts
- 10. JUNGKOOK FOR CHANEL BEAUTY 144K posts
- 11. Trey Songz N/A
- 12. LING AVATAR FIRE AND ASH TH 488K posts
- 13. #หลิงหลิงอวตารอัคนีและธุลีดิน 509K posts
- 14. Dolly 14.9K posts
- 15. Doug Dimmadome 15.6K posts
- 16. #thursdaymotivation 2,388 posts
- 17. Ukraine and Israel 6,406 posts
- 18. Confessions 4,928 posts
- 19. Code Pink 3,432 posts
- 20. Lupin 3,962 posts